Oncoscience
Suppression of vascular endothelial growth factor expression in breast cancer cells by microRNA-125b-mediated attenuation of serum amyloid A activating factor-1 level
Alpana Ray1 and Bimal K. Ray1
1 Department of Veterinary Pathobiology, University of Missouri, Columbia, Missouri
Correspondence to: Bimal K. Ray, email: [email protected]
Keywords: Breast cancer; SAF-1, miR-125b; transcriptional regulation; VEGF expression
Received: December 05, 2018
Accepted: April 5, 2019
Published: July 2, 2019
Copyright: Ray et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Increased level of an inflammation-responsive transcription factor called serum amyloid A-activating factor (SAF-1) has been linked to the pathogenesis in human breast cancer. SAF-1 is found to promote vascular endothelial growth factor (VEGF) expression in breast carcinoma cells and boost angiogenesis. In an effort to develop a cellular mechanism to control VEGF expression, we sought to limit SAF-1 activity in breast cancer cells. We report here several targets within the SAF-1 mRNA for binding of microRNA-125b (miR-125b) and we show that VEGF expression is reduced in breast cancer cells when SAF-1 level is reduced with the microRNA action. Within the 3´ un-translated region (UTR) of SAF-1 transcript, we have identified four highly conserved miR-125b responsive elements. We show that these miR-125b binding sites mediate repression of SAF-1 by miR-125b. Ectopic expression of miR-125b in nonmetastatic and metastatic breast cancer cells repressed SAF-1-mediated activity on VEGF promoter function and inhibited cancer cell migration and invasion potentials in vitro. Together, these results suggest that termination of SAF-1 function by miR-125b could be developed as a potential anti-VEGF and anti-angiogenic agent, which has high clinical relevance.
INTRODUCTION
Enhanced vascular endothelial growth factor (VEGF) expression is an important event in the growth and metastatic spread of breast cancer [1, 2]. VEGF, as an angiogenic factor, promotes new blood vessel formation. This process called angiogenesis plays a central role in the growth and progression of solid tumors by supplying nutrients, growth factors and oxygen and eventually assists in metastasis [3, 4]. Over-expression of VEGF in tumors primarily occurs due to transcriptional induction of VEGF. VEGF expression is induced by many factors, including cytokines, growth factors, hypoxia, differentiation, oncogenic transformation, which suggests for the presence of multiple routes to VEGF mRNA synthesis [5-7]. Since tumor cells are quite adept at acquiring new alternative processes to circumvent surrounding environmental pressures, identification of all regulatory pathways in tumor cells controlling VEGF expression is important. An inflammation-responsive transcription factor, called serum amyloid A-activating factor 1 (SAF-1) has been identified as a transcriptional regulator of VEGF during angiogenic switch in arthritic animals [ 8]. Later we have shown that SAF-1 is abundantly present in human breast cancer tissues and up-regulates VEGF expression in metastatic breast cancer cells [9].
High level of SAF-1 in breast cancer tissues and cells could also have additional implication for its role in cancer development and progression. For example, SAF-1 has been shown to regulate expression of several genes that play important role in cell invasion, cell proliferation and blood vessel remodeling. SAF-1 regulates several matrix metalloproteinases (MMPs) including MMP-1, MMP-9 and MMP-14 which are involved in extracellular matrix degradation and thus implicated in blood vessel growth and metastasis [10-12]. Human orthologue of SAF-1 regulates endothelial nitric-oxide synthase that plays quintessential role in remodeling and regulation of blood vessel [13] and ephrin-B2, a transmembrane ligand for Eph receptors that are implicated in angiogenic remodeling [14]. Human SAF-1 is shown to regulate c-myc proto-oncogene [15] and drive tumor specific expression of PPARγ1 [16], overexpression of which correlates with breast cancer metastasis. Regulatory function of SAF-1 is mediated by its activity as a transcription factor. SAF-1 has a Cys2-His2-type zinc finger that is common to many other regulatory transcription factors such as Sp 1 and KLF family of transcription factors, just to name a few, and it is activated during inflammation [17-21]. Given the involvement of SAF-1 in regulating genes controlling tumor angiogenesis, cellular invasion, tumor cell proliferation and differentiation, and our previous finding of overexpression of SAF-1 in breast cancer [9], we aimed at developing a new approach to modulate SAF-1 by pharmacological treatment which would be an effective and promising method for cancer therapy. Indeed, the feasibility of this approach is validated by the observation of synthetic siRNA-mediated suppression of SAF-1 activity [9]. Availability of a natural RNA inhibitor would be more preferable for effective suppression of cellular SAF-1 gene. Here we report that a naturally occurring micro RNA, miR-125b, has the potential of regulating SAF-1 expression.
MicroRNAs (miRNAs) are endogenous small noncoding RNAs (20–23 nucleotides) and are known to regulate many biological processes, including development, cellular proliferation, apoptosis and differentiation [22]. In the cytoplasm, miRNAs negatively regulate gene expression at the post-transcriptional level by base pairing to the 3′ un-translated region (UTR) of target messenger RNAs and cause either mRNA degradation or translational arrest or both [22]. The miRNAs are found to be aberrantly expressed or mutated in cancer, suggesting that they may function as tumor suppressors or oncogenes depending on their expression status [reviewed in 23]. When miRNAs are over-expressed and inactivate tumor promoting molecules such as growth factors, they function as tumor suppressor [23]. Conversely, mutation in miRNAs may have an opposite effect. For example, when myc gene is translocated in B-cell leukaemia within the miR-142 locus, it causes mutation of this miRNA. Such a translocation results in an increased expression of myc and suppression of miR-142 expression. Thus, mutation of miR-142 has an oncogenic effect in B-cell leukaemia [23]. Among the different miRNA families, miR-125 family has been found to be associated with a variety of carcinomas [24]. In breast cancer, high level of expression of miR-125b has been shown to cause down-regulation of ERBB2 (HER2) and ERBB3 (HER3), and thereby suppression of tumor growth [25].
Since miR-125b has been shown to target VEGF expression in hepatocellular carcinoma [26], and our research has shown that VEGF expression is up-regulated by SAF-1 in breast cancer [9], we investigated whether SAF-1 could be a target of miR-125b. In the present study, we show that within the 3´UTR of human SAF-1 mRNA, four highly conserved miR-125b-responsive elements are present, in which three are adjacently present. Here, we also show that the repressive effect of miR-125b on SAF-1 is mostly mediated by these clustered miR125b-responsive elements. Furthermore, here we show that ectopic overexpression of miR-125b in breast cancer cells reduces SAF-1 expression leading to decrease of VEGF level and consequential diminution of cancer cell migration and invasion. In correlation with previous microarray studies indicating decreased miR-125b expression in breast cancer tissues and miR-125b to be one of the most consistently deregulated miRNAs in breast cancer [27, 28], we report here that miR-125b is downregulated in breast cancer cells.
Since high level of SAF-1 in breast cancer is linked to angiogenesis, cancer cell proliferation and metastasis, a new approach to curb the activity of SAF-1 using microRNA may provide a new option in pharmacological treatment for breast cancer therapy.
RESULTS
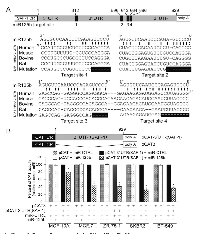
Multiple miR-125b responsive elements in human SAF-1 mRNA
Computer analysis to search for microRNA target recognition sites in the SAF-1 mRNA revealed four highly conserved sequence elements comprised of complementary core segment or the “seed” region of miR-125b binding element within the 929 nucleotides long 3´UTR (Figure 1A). The first miR-125b element (target site 1) is present about 300 nucleotides upstream of three clustered miR-125b elements (target sites 2-4). To determine if there is an interaction with miR-125b, the entire 3´-UTR of human SAF-1 was ligated at the 3´end of the translational unit encoding chloramphenicol acetyl transferase (CAT) in the pCAT3 reporter plasmid. Expression of this reporter plasmid, pCAT-3´UTR (SAF1), in response to ectopic overexpression of miR-125b RNA was found to be substantially decreased in normal breast epithelial MCF-10A cells and in several breast cancer cells including ER positive MCF-7 and ZR-75-1 and ER-negative SKBR3 and BT-549 cells (Figure 1B). However, ectopic overexpression of scrambled miR RNA (miR-CTRL) did not decrease expression of the reporter plasmid, pCAT-3´UTR (SAF1), expression. Furthermore, pCAT3 reporter vector, that did not contain the 3´-UTR region of SAF-1, was not inhibited in response to overexpression of miR-125b RNA or scrambled RNA. Together these results suggested that the 3´-UTR of human SAF-1 mRNA is a potential target for repression by miR-125b.
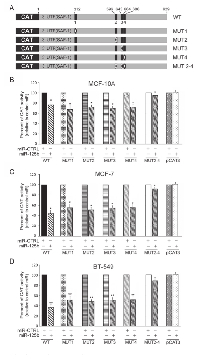
Mutational analysis of miR-125b-responsive elements
To determine the individual contribution among the four miR-125b target sites in SAF-1 mRNA 3´UTR, reporter plasmids containing deletion mutation of each of these four target sites, as indicated in Fig. 1A, were constructed. Four mutant plasmids with a single site mutated, referred to as MUT1 to MUT4, and one mutant with 3 sites mutated, MUT2-4 (Figure 2A), were used in the mutation analysis. These five reporter constructs with various mutated SAF-1 3´UTR were transfected into normal breast epithelial and breast cancer cells. Results are shown in Figures 2B-D. Deletion of a single element (MUT1, MUT2, MUT3 or MUT4) reduced but did not drastically eliminate miR-125b-mediated repression of pCAT-3´UTR (SAF1) reporter in MCF-10A or MCF-7 and BT-549 breast cancer cells (Figure 2B-D). However, when three clustered target sites of SAF-1 3´-UTR were mutated together in MUT2-4, there was almost no repressive effect of miR-125b. These results suggested that the clustered miR-125b target sites are the principal elements within the human SAF-1 3´-UTR that mediate repression by miR-125b.
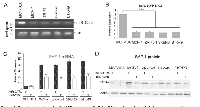
Impact of miR-125b overexpression on SAF-1 abundance in breast cancer cells
We noticed that in MCF-10A cells, the repressive effect of miR-125b is of much smaller magnitude as compared to that observed in the breast cancer cells (Figures 1 and 2). To investigate this phenomenon endogenous miR-125b expression levels in MCF-10A, MCF-7, ZR-75-1, SKBR3 and BT-549 cells were examined by semi-quantitative RT-PCR (Figure 3A) and qRT-PCR analysis (Figure 3B).These analyses clearly revealed that MCF-10A cells express miR-125b RNA at a much higher level than all breast cancer cells that were examined. Ectopic or forced overexpression of miR-125b RNA in these breast cancer cells substantially reduced SAF-1 mRNA (Figure 3C) and SAF-1 protein (Figure 3D) levels, which correlated with the 3’ UTR-reporter assays as seen in Figures 1 and 2.
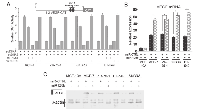
Overexpression of miR-125b down-regulates SAF-1-mediated VEGF expression
As SAF-1 is a transcriptional inducer of VEGF [9], we examined the effect of miR-125b mediated repression of SAF-1 on VEGF expression. The transcription from 1.2VEGF-CAT reporter, which is dependent on the availability of SAF-1 [ 9], was down-regulated by miR-125b but not by control miRNA (Figure 4A). In correlation, ectopic expression of miR-125b RNA lowered VEGF mRNA (Figure 4B) and VEGF protein (Figure 4C) levels in the transfected breast cancer cells. In MCF-10A cells this response was minimum, which correlated with earlier results (Figure 3A-D).
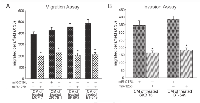
miR-125b impairs cell migration and invasive potentials of breast cancer cells
All tumors must undergo angiogenesis to acquire nutrients for continued growth and metastatic spread. VEGF accounts for a cell migration of vascular endothelial cells and cellular invasion of tumor cells in cancer [29]. To verify if down-regulation of SAF-1 by miR-125b and subsequent reduction in the VEGF level has any measurable effect on angiogenic processes, we examined cellular migration of vascular endothelial cells (HUVEC). HUVEC were plated in trans-well chambers, incubated in presence of culture medium fortified with CM from breast cancer cells transfected with pre-mir-125b RNA, and observed for their ability to migrate through the membrane. Increase of miR-125b level resulted in ~ 50% lowered migration rate of cells as compared to scrambled pre-miR RNA transfected cells (Figure 5A). Invasiveness, requiring both cell motility and ability to advance through a solid and proteinaceous extracellular matrix like Matrigel, is thought to more directly assess cell metastatic potential. MCF-7 and ZR-75-1 breast cancer cells are regarded as noninvasive breast cancer cells, while SKBR3 and BT-549 cells exhibit invasive characteristics. Overexpression of miR-125b RNA, substantially reduced cellular invasive phenotype of both SKBR3 and BT-549 cells as compared to control pre-miR oligonucleotide (Figure 5B).
DISCUSSION
Phosphorylation caused by cytokine- and growth factor-mediated signal transduction events plays a key role in the induction of transcriptional activity and DNA-binding activity of SAF-1 [19-21]. Increased SAF-1 activity is linked to pathogenesis of diseases including breast cancer [8-11, 30, 31]. While much is known about the activation of SAF-1, relatively little is known about its down-regulation or control in the cellular environment. Understanding of such a regulatory process would be useful in breast cancer therapy. Therefore, to identify a possible manner by which SAF-1 could be controlled intracellularly, we explored the possibility of microRNA mediated regulation of SAF-1. Our findings, described here, provide strong evidence that SAF-1 transcription factor is regulated by the microRNA miR-125b. The conclusion that miR-125b suppresses SAF-1 is supported by (i) demonstration of miR-125b elements in SAF-1 3´-UTR, mutation of which abolishes all regulatory effects of miR-125b on SAF-1 (Figures 1 and 2). We further show that (ii) overexpression of miR-125b causes reduction of SAF-1 mRNA level and protein expression in breast cancer cells (Figure 3), (iii) suppression of VEGF expression (Figure 4) and (iv) reduction of cell migration and (v) cell invasion in different types of breast cancer cells. (Figure 5)
The miR-125b target elements in the 3´UTR of SAF-1 mRNA are highly conserved across many species from human to rat (Figure 1) suggesting the possibility that these regulatory elements may play an important role in maintaining homeostasis in multiple species. We identified four elements among which three are adjacently present. Although, each of the miR-125b elements is similarly potent in mediating repression, simultaneous mutation of the three adjacent elements nearly abolished all repressive effects of miR-125b. Thus the three clustered elements appear to be the principal elements that mediate repression by miR-125b and necessary for efficient down-regulation of SAF-1.
The observed reduction in SAF-1 protein level in cancer cells following ectopic overexpression of miR-125b (Figure 3) can be reasonably attributed to miRNA effects on SAF-1 transcript stability and translation inhibition, as a previous report indicates that miR-125b can bifunctionally mediate both transcript decay and translational inhibition [32]. It will be interesting to find out whether relative positioning of the four miR-125b target elements in SAF-1 3´UTR would affect mRNA stability or translation or both.
Down regulation of miR-125b has been reported in primary breast cancers and metastatic breast tumors tissues [27, 33, 34]. We have also found much lower levels of miR-125b RNA in breast cancer cells in comparison to normal MCF-10A cells (Figure 3). Higher level of endogenous miR-125b RNA pool in MCF-10A cells can explain why SAF-1 mRNA and SAF-1 protein levels are so low in these cells as compared to those in breast cancer cells. This finding also explains the results in Figure 1B, 2C and 2D and suggests that the presence of already high level of endogenous miR-125b in MCF-10A cells most likely significantly suppresses most of SAF-1 level and thus further ectopic expression of miR-125b probably has less suppressive effect on SAF-1 in these cells. In contrary, in breast cancer cells, since endogenous level of miR-125b is low, there is very little suppression of SAF-1 level and thus further ectopic expression of miR-125b significantly suppresses SAF-1. This explains high level of inhibition of CAT activity by miR-125b in cancer cells which is seen Figures 1 and 2.
Our findings show that inhibition of endogenous SAF-1 activity by miR-125b action results in the significant inhibition of VEGF promoter function in the breast cancer cells (Figure 4) where the SAF-1 activity is much higher compared to normal cells [9]. This suggests a possible functional role of miR-125b in angiogenesis. Our previous studies have shown that in normal breast epithelial cells, MCF-10A, the CAT reporter expression in cells transfected with 1.2 VEGF-CAT reporter is very low and ectopic overexpression of SAF-1 in MCF-10A cells has minimal effect [35]. This suggested that in MCF-10A, high pool of endogenous miR-125b is able to sufficiently repress SAF-1 mRNA. In contrary, the breast cancer cells exhibit higher reduction of CAF activity under same condition suggesting lower level of endogenous miR-125b in these cells.
The microRNA mediated regulations are being increasingly implicated in tumorigenesis. Normally, miR-125b is expressed in many tissue types but its level is seen to be induced during neural differentiation, neurogenesis and during in vitro retinoic acid treatment of cells. However, the role of miR-125b in tumorigenesis is somewhat complex due to its diverse functions in different cell contexts [24]. In breast cancer and hepatocellular carcinoma, miR-125b expression becomes lower and such low expression is shown to be consequential for increased expressions of a number of cancer-associated genes that support tumorigenesis. These findings suggested miR-125b to be a tumor suppressor [25, 27, 28, 36-38]. However, expression of miR-125b is seen to be elevated in prostate cancer [39] and in a range of human leukemias [40-42] and ectopic expression of miR-125b is seen to favor stimulate cell proliferation and tumorigenicity [39] and cause highly invasive myeloid leukemia in the animals [43]. These diverse observations suggest that the role of miR-125b in cancer is dependent on the distinct pathophysiological conditions and the target molecules for the microRNA in different disease conditions. For therapeutic purposes, a careful selection of microRNA target may provide a desirable outcome.
In the present study, we observed that miR-125b overexpression reduces SAF-1 level. The effect of lower SAF-1 level, translated into a drop in VEGF expression and lowering of the tumorigenic potentials of breast cancer cells.
In summary, we report that SAF-1 is a direct target of miR-125b microRNA and reduced expression of miR-125b in breast cancer cells, partially accounts for upregulation of SAF-1 and VEGF in breast cancer. Regulation of miR-125b may have further implications in the complex events of tumorigenesis.
METHODS
Cell lines
MCF-10A, MCF-7, ZR-75-1, SKBR3, BT-549, and HUVEC-CS cells were obtained from American Type Culture Collection (ATCC), cultured and stored following ATCC protocol of authentication by STR analysis. The cells were maintained in Dulbecco modified Eagle medium (DMEM)-high glucose medium supplemented with 10% fetal bovine serum. For harvesting conditioned medium (CM) we followed a method that was described earlier [35]. Briefly, the cells were first grown in DMEM containing 7% FCS for 24 h. Next, the culture medium was replaced with DMEM containing 0.5% FCS, transfected with either miR-125b or miR-CTRL RNA and the cells were grown for an additional 48 h, after which the medium was collected, centrifuged at 1,000 x g, and stored at -80 0 C for further use.
Plasmid constructs, transfection and CAT reporter assay
The reporter plasmid 1.2VEGF-CAT3 was described earlier [9].The pCAT-3´UTR (SAF1) was prepared by ligating 929 base pairs of human SAF-1 cDNA sequences representing entire 3´un-translated region (3´UTR) [44, 45] to the downstream of the coding sequence of chloramphenicol acetyl transferase (CAT) gene at the Xba I site in the pCAT3 promoter vector (Promega Corporation). The mutant reporters were constructed by megaprimer PCR amplification in which the individual miR-125b elements in the 3´ UTR of SAF-1 were mutated using mutated PCR primers. Cells were transfected using lipofectamine 2000 (Invitrogen) following manufacturer’s protocol. For CAT assay, cells were transfected by adding reporter plasmid DNA together with pSVβ-gal (Promega Corporation) plasmid DNA as described [11]. The pSVβ-gal DNA was used to monitor the efficiency of transfection and to normalize cell extracts used for CAT assay. To study the biological effects of miR-125b on breast cancer cells, cells were transfected with 50 nM of premiR-125b RNA (Invitrogen/Ambion; PM10148) or 50 nM of negative control (pre-miR-CTRL, Invitrogen/Ambion) molecules following the manufacturer’s protocol.
RNA isolation, semi-quantitative and quantitative RT-PCR analysis
The total RNA was isolated using a RNA isolation kit (Qiagen). Expression of miR-125b was assessed by semiquantitative RT-PCR, using a small RNA specific RT-PCR detection kit (Quantimir RT; Systems Biosciences) with the universal reverse primer and a human miR-125b specific forward primer (5´-TCCCTGAGTCCCTAACTTGT-3´) by initial denaturation at 94 °C for 2 min, 22 cycles of 94 °C for 30s, 58 °C for 30s, 72 °C for 30s and a final extension for 5 min at 72 °C. The products were run in an agarose gel. For quantitative real-time PCR, the miRNA specific TaqMan MicroRNA Assay kits (Applied Biosystems) for miR-125b, SAF-1, VEGF and GAPDH were used by following manufacturer’s protocol. Expression levels were normalized against two reference sequences, RNU44, RNU48 and/or GAPDH and relative changes were calculated. Experiments were run in triplicates.
Western blot analysis
Cell extracts (70 µg) were fractionated using sodium dodecyl sulfate (SDS)-11% polyacrylamide gel electrophoresis (PAGE) and electroblotted onto PVDF membrane. The rabbit anti-VEGF and anti-β-actin antibodies were obtained from Santa Cruz Biotechnology. The rabbit anti-SAF-1 antibody, as described [10], was used at 1:5000 dilution. Bands were detected using a chemiluminescence detection kit (Amersham Biosciences).
Cell migration and Invasion Assays
HUVEC migration assay was performed as described earlier (9, 35). Cells were trypsinized, pelleted and resuspended in medium without serum or growth factors. For migration assay, 1 x 105 HUVEC cells in 0.5 ml medium were added onto 24-well transwell Boyden chamber with an 8-µm pore polycarbonate membrane. For invasion assay, 1 x 105 SKBR3 and BT-549 breast cancer cells were placed onto the transwells which were precoated with 20 µg of Matrigel (BD Biosciences). For both cell migration and invasion assays, the transwells were then placed over bottom wells filled with conditioned medium (CM) of either miR-125b or miR-control (CTRL) RNA transfected breast cancer cells. After 24 h of incubation, HUVEC cells that did not migrate or cancer cells that did not invade through matrigel and remained on the insert top layers were removed with cotton swab and cells on the membrane lower surface, representing migrated and invaded cells, were fixed with 100% methanol for 2 min followed by staining with 1% toluidine blue solution. Inserts were washed several times before air drying; membranes were photographed and cell counts were determined after totaling five random fields. All assays were replicated three times.
Statistics
To compare multiple sets of data, a one-way analysis of variance (ANOVA) with post hoc Fisher’s least significant difference test was used. For paired data sets, a two-tailed t test was used. Values of P < 0.05 were considered to represent a significant difference.
ABBREVIATIONS
SAF-1, serum amyloid A-activating factor-1; VEGF, vascular endothelial growth factor; miR-125b, micro RNA-125b; CTRL RNA, negative control RNA; UTR, un-translated region; siRNA, short interfering RNA; RT-PCR, reverse transcription polymerase chain reaction; qRT-PCR, quantitative RT-PCR; EMSA electrophoretic mobility shift assay; PAGE, polyacrylamide gel electrophoresis; CAT, chloramphenicol acetyl transferase.
AUTHOR CONTRIBUTIONS
AR and BKR designed the study, performed experiments, acquired data, interpreted the results and prepared the manuscript. Both authors approved the final content for journal submission and publication
ACKNOWLEDGMENTS
We thank members of the Cancer Research Program for discussions and comments during the study.
CONFLICTS OF INTEREST
None of the authors declare any conflict of interest.
FUNDING
This study was supported by the University of Missouri, College of Veterinary Medicine.
- 1. Expression of vascular endothelial growth factor, its receptor, and other angiogenic factors in human breast cancer. Cancer Res. 1996; 56:2013–16. http://cancerres.aacrjournals.org/content/56/9/2013.long . [PubMed].
- 2. Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in breast cancer. Hum Pathol. 1995; 26:86–91. https://doi.org/10.1016/0046-8177(95)90119-1. [PubMed].
- 3. Role of vascular endothelial growth factor in physiologic and pathologic angiogenesis: therapeutic implications. Semin Oncol. 2002 (Suppl 16); 29:10–14. https://doi.org/10.1016/S0093-7754(02)70064-X. [PubMed].
- 4. Increased angiogenesis and lymphangiogenesis in inflammatory versus noninflammatory breast cancer by real-time reverse transcriptase-PCR gene expression quantification. Clin Cancer Res. 2004; 10:7965–71. https://doi.org/10.1158/1078-0432.CCR-04-0063. [PubMed].
- 5. The biology of vascular endothelial growth factor. Endocr Rev. 1997; 18:4–25. https://doi.org/10.1210/edrv.18.1.0287. [PubMed].
- 6. Clinical translation of angiogenesis inhibitors. Nat Rev Cancer. 2002; 2:727–39. https://doi.org/10.1038/nrc905. [PubMed].
- 7. Protein kinases and the hypoxia-inducible factor-1, two switches in angiogenesis. Curr Pharm Des. 2003; 9:531–41. https://doi.org/10.2174/1381612033391469. [PubMed].
- 8. Vascular endothelial growth factor expression in arthritic joint is regulated by SAF-1 transcription factor. J Immunol. 2007; 178:1774–82. https://doi.org/10.4049/jimmunol.178.3.1774. [PubMed].
- 9. Control of VEGF expression in triple-negative breast carcinoma cells by suppression of SAF-1 transcription factor activity. Mol Cancer Res. 2011; 9:1030–41. https://doi.org/10.1158/1541-7786.mcr-10-0598. [PubMed].
- 10. Induction of matrix metalloproteinase 1 gene expression is regulated by inflammation-responsive transcription factor SAF-1 in osteoarthritis. Arthritis Rheum. 2003; 48:134–45. https://doi.org/10.1002/art.10706. [PubMed].
- 11. Induction of the MMP-14 gene in macrophages of the atherosclerotic plaque: role of SAF-1 in the induction process. Circ Res. 2004; 95:1082–90. https://doi.org/10.1161/01.RES.0000150046.48115.80. [PubMed].
- 12. Transcriptional induction of matrix metalloproteinase-9 in the chondrocyte and synoviocyte cells is regulated via a novel mechanism: evidence for functional cooperation between serum amyloid A-activating factor-1 and AP-1. J Immunol. 2005; 175:4039–48. https://doi.org/10.4049/jimmunol.175.6.4039. [PubMed].
- 13. Characterization of the human endothelial nitric-oxide synthase promoter. J Biol Chem. 1999; 274:3076–93. https://doi.org/10.1074/jbc.274.5.3076. [PubMed].
- 14. Characterization of the murine Ephrin-B2 promoter. Gene. 2009; 437:54–59. https://doi.org/10.1016/j.gene.2009.02.017. [PubMed].
- 15. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc Natl Acad Sci USA. 1992; 89:7452–56. https://doi.org/10.1073/pnas.89.16.7452. [PubMed].
- 16. MAZ drives tumor-specific expression of PPAR gamma 1 in breast cancer cells. Breast Cancer Res Treat. 2008; 111:103–11. https://doi.org/10.1007/s10549-007-9765-7. [PubMed].
- 17. Activation of Sp1 and its functional co-operation with serum amyloid A-activating sequence binding factor in synoviocyte cells trigger synergistic action of interleukin-1 and interleukin-6 in serum amyloid A gene expression. J Biol Chem. 1999; 274:4300–08. https://doi.org/10.1074/jbc.274.7.4300. [PubMed].
- 18. Involvement of an SAF-like transcription factor in the activation of serum amyloid A gene in monocyte/macrophage cells by lipopolysaccharide. Biochemistry. 1997; 36:4662–68. https://doi.org/10.1021/bi9624595. [PubMed].
- 19. Activation of transcription factor SAF involves its phosphorylation by protein kinase C. J Biol Chem. 2000; 275:39727–33. https://doi.org/10.1074/jbc.M007907200. [PubMed].
- 20. Cytokine-responsive induction of SAF-1 activity is mediated by a mitogen-activated protein kinase signaling pathway. Mol Cell Biol. 2002; 22:1027–35. https://doi.org/10.1128/MCB.22.4.1027-1035.2002. [PubMed].
- 21. Protein kinase A signaling pathway regulates transcriptional activity of SAF-1 by unmasking its DNA-binding domains. J Biol Chem. 2003; 278:22586–95. https://doi.org/10.1074/jbc.M300705200. [PubMed].
- 22. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–97. https://doi.org/10.1016/S0092-8674(04)00045-5. [PubMed].
- 23. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006; 6:259–69. https://doi.org/10.1038/nrc1840. [PubMed].
- 24. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013; 6:6. https://doi.org/10.1186/1756-8722-6-6. [PubMed].
- 25. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007; 282:1479–86. https://doi.org/10.1074/jbc.M609383200. [PubMed].
- 26. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One. 2012; 7:e40169. https://doi.org/10.1371/journal.pone.0040169. [PubMed].
- 27. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005; 65:7065–70. https://doi.org/10.1158/0008-5472.CAN-05-1783. [PubMed].
- 28. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006; 5:24. https://doi.org/10.1186/1476-4598-5-24. [PubMed].
- 29. Ray A, Kumar D, Shakya A, Brown CR, Cook JL, Ray BK. Serum amyloid A-activating factor-1 (SAF-1) transgenic mice are prone to develop a severe form of inflammation-induced arthritis. J Immunol. 2004; 173:4684-91. https://doi.org/10.4049/jimmunol.173.7.4684. [PubMed].
- 30. Inflammation-responsive transcription factor SAF-1 activity is linked to the development of amyloid A amyloidosis. J Immunol. 2006; 177:2601–09. https://doi.org/10.4049/jimmunol.177.4.2601. [PubMed].
- 31. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005; 25:9198–208. https://doi.org/10.1128/MCB.25.21.9198-9208.2005. [PubMed].
- 32. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009; 219:214–21. https://doi.org/10.1002/path.2586. [PubMed].
- 33. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007; 8:R214. https://doi.org/10.1186/gb-2007-8-10-r214. [PubMed].
- 34. Fighting cancer by attacking its blood supply. Sci Am. 1996; 275:150–54. https://doi.org/10.1038/scientificamerican0996-150. [PubMed].
- 35. Loss of epigenetic Kruppel-like factor 4 histone deacetylase (KLF-4-HDAC)-mediated transcriptional suppression is crucial in increasing vascular endothelial growth factor (VEGF) expression in breast cancer. J Biol Chem. 2013; 288:27232–42. https://doi.org/10.1074/jbc.M113.481184. [PubMed].
- 36. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011; 71:3552–62. https://doi.org/10.1158/0008-5472.CAN-10-2435. [PubMed].
- 37. A short hairpin DNA analogous to miR-125b inhibits C-Raf expression, proliferation, and survival of breast cancer cells.[Erratum appears in Mol Cancer Res. 2010 Jun;8(6):935 Note: Radziwil, Gerald [corrected to Radziwill, Gerald]]. Mol Cancer Res. 2009; 7:1635–44. https://doi.org/10.1158/1541-7786.MCR-09-0043. [PubMed].
- 38. miR-125b targets ARID3B in breast cancer cells. Cell Struct Funct. 2012; 37:27–38. https://doi.org/10.1247/csf.11025. [PubMed].
- 39. Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, deVere White RW. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007; 104:19983-88. https://doi.org/10.1073/pnas.0706641104. [PubMed].
- 40. Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, Lippert E, Talmant P, Lafage-Pochitaloff M, Leroux D, Gervais C, Viguié F, Lai JL, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008; 205:2499-506. https://doi.org/10.1084/jem.20080285. [PubMed].
- 41. Eμ/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia. 2011; 25:1849–56. https://doi.org/10.1038/leu.2011.166. [PubMed].
- 42. Klusmann JH, Li Z, Böhmer K, Maroz A, Koch ML, Emmrich S, Godinho FJ, Orkin SH, Reinhardt D. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010; 24:478-90. https://doi.org/10.1101/gad.1856210. [PubMed].
- 43. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci USA. 2012; 109:4233–38. https://doi.org/10.1073/pnas.1200677109. [PubMed].
- 44. SAF-3, a novel splice variant of the SAF-1/MAZ/Pur-1 family, is expressed during inflammation. FEBS J. 2009; 276:4276–86. https://doi.org/10.1111/j.1742-4658.2009.07136.x. [PubMed].
- 45. Isolation and functional characterization of cDNA of serum amyloid A-activating factor that binds to the serum amyloid A promoter. Mol Cell Biol. 1998; 18:7327–35. https://doi.org/10.1128/MCB.18.12.7327. [PubMed].
Last Modified: 2019-08-23 14:24:45 EDT
