Oncoscience
Abstract | PDF | Full Text | Supplementary Materials | How to Cite
https://doi.org/10.18632/oncoscience.37
A novel cohort of cancer-testis biomarker genes revealed through meta-analysis of clinical data sets.
Stephen J. Sammut1,*, Julia Feichtinger2,3,*, Nicholas Stuart1, Jane A. Wakeman4, Lee Larcombe4, Ramsay J. McFarlane4,5
1 School of Medical Sciences, Bangor University, Bangor, UK
2 Institute for Knowledge Discovery, Graz University of Technology, Austria
3 Core Facility Bioinformatics, Austrian Centre of Industrial Biotechnology, Austria
4 North West Cancer Research Institute, Bangor University, Bangor, UK
5 NISCHR Cancer Genetics Biomedical Research Unit
* These authors made an equal contribution to this work
Correspondence to: Ramsay J. McFarlane, email: [email protected]
Keywords: Cancer/testis antigens; cancer biomarkers; meiCT; gene expression; oncogenes; meiosis
Received: April 24, 2014
Accepted: May 06, 2014
Published: May 06, 2014
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT:
The identification of cancer-specific biomolecules is of fundamental importance to the development of diagnostic and/or prognostic markers, which may also serve as therapeutic targets. Some antigenic proteins are only normally present in male gametogenic tissues in the testis and not in normal somatic cells. When these proteins are aberrantly produced in cancer they are referred to as cancer/testis (CT) antigens (CTAs). Some CTA genes have been proven to encode immunogenic proteins that have been used as successful immunotherapy targets for various forms of cancer and have been implicated as drug targets. Here, a targeted in silico analysis of cancer expressed sequence tag (EST) data sets resulted in the identification of a significant number of novel CT genes. The expression profiles of these genes were validated in a range of normal and cancerous cell types. Subsequent meta-analysis of gene expression microarray data sets demonstrates that these genes are clinically relevant as cancer-specific biomarkers, which could pave the way for the discovery of new therapies and/or diagnostic/prognostic monitoring technologies.
INTRODUCTION
Achieving effective treatments for cancers is more difficult once the disease has reached the metastatic stage. This, in combination with the trend towards personalised approaches to cancer medicine means there is an increasing need to identify and develop cancer-specific biomarkers that can be employed in the development of early, pre-metastatic diagnostic and treatment strategies [1-7]. Additionally, advances in tumour immunology have reignited interest in cancer immunotherapeutics, such as cancer vaccines, adoptive therapeutics and targeted drug delivery using antibody-drug conjugates [8-17].
Cancer/testis antigens (CTAs) have emerged as a group of proteins that have significant immunogenic and cancer-specific potential [18-27]. Bona fide CTA genes are defined by having expression restricted to the testes in normal adult males, but are also aberrantly activated in cancers of either gender [18-27]. CTA genes are of importance for two fundamentally distinct reasons. Firstly, there is an immunological barrier, known as the blood-testis barrier, which generates an immunological privilege within the testis that is enforced via a number of pathways [28, 29]; consequently, testis antigens that normally reside in an immunologically privileged setting are capable of eliciting an autologous immune response in the peripheral blood/tissues. Thus, CTAs can serve as immunologically restricted cancer-specific antigens, making them exceptionally attractive as diagnostic, prognostic and therapeutic biomarkers/targets, the targeting of which should not induce deleterious side effects to non-cancerous somatic tissue. For example, the activation of a specific cohort of such genes has been correlated with more aggressive lung cancers [30]. Additionally, use of an immunohistochemical approach in non-small cell lung cancers revealed a correlation between survival and the presence of known CTAs [31]. Of note, the presence of the CTA NY-ESO-1 correlated with an increased response to and benefit from neoadjuvant, and adjuvant chemotherapy respectively [31]. NY-ESO-1 is one of the most immunogenic CTAs and has been used as a successful target for adoptive therapy in the treatment of malignant melanoma [32].
Secondly, genes whose products normally serve to drive meiotic chromosome dynamics, germ cell regulation and gametogenic differentiation may have powerful oncogenic transforming activity if aberrantly expressed in non-germline, somatic tissues; a possibility that remains largely unexplored. The aberrant activation of these genes may confer biological processes that are advantageous to cancer cells but at the same time may be open to exploitation by therapeutic targeting (for examples, see 33,34). Indeed, increasing evidence indicates that a soma-to-germline transition could be a potential broad-spectrum oncogenic driver [35-39].
CTA genes have been classified based on expression profiles in normal tissues. These groups are: testis-restricted, testis-selective, testis/brain [or central nervous system (CNS) tissue]-restricted and testis/brain (CNS tissue) selective respectively [40]. Most of the known CTAs are encoded for by genes on the X chromosome (X-CT genes)[18, 26, 40], and many belong to large paralogous gene families (for example, see 41). Previously, meiosis-specific genes have been identified as CTA genes (for example, see 42,43), but recently a more systematic study described an extensive set of putative meiosis-specific genes, meiCT (or meiCTA) genes, as cancer/testis (CT) genes [44]; many of these are autosomally encoded, fitting with the fact that the X chromosome becomes transcriptionally silenced in mammalian male meiosis [45, 46].
Given the strict cancer specificity of CTAs, the identification of new CT genes has exceptional therapeutic and biomarker potential. In this study, we describe a novel sub-category of meiCT genes, which have clinical importance, as demonstrated through meta-analysis of a clinically-relevant gene expression microarray data sets.
RESULTS
A bioinformatics pipeline was previously established to identify putative human meiosis-specific genes that could potentially encode CTAs. This was based initially on a cohort of mouse genes predicted to be associated specifically with meiosis and spermatocyte development [44]. High stringency human orthologue identification and filtering for mitotic expression resulted in 375 human genes which were potentially testis spermatocyte /meiosis-specific [44]. These genes were then evaluated using an EST analysis pipeline based on the complete Unigene database [44]. Briefly, if a candidate gene was represented in a non-testis/non-central nervous system (CNS) normal tissue EST library, then it was excluded. The remaining genes were assessed further to see if they were represented in cancer EST libraries. From the original 375 potential testis-specific genes the EST analysis identified 177 candidate genes, of which 9 were testis-restricted, but also gave a positive cancer EST signature (class 1); 75 were testis-restricted, with no cancer EST signature (class 2); 21 were testis/CNS-restricted, with a positive EST signature (class 3) and 72 were testis/CNS-restricted, with no cancer EST signature (class 4).
We have previously defined the meiCT genes based on validation and gene expression microarray analyses of the class 1-3 genes [44]. Within the initial class 1-3 predicted gene sets RT-PCR validation revealed that a number were actually expressed in extensive somatic tissues [44]. Given this, we re-analysed our predicted class 4 genes using an updated CancerEST pipeline [47] and from this we identified 54 putative class 4 genes, those with expression signatures only in the testis and CNS of healthy tissue (Supplementary Table).
The gene expression profiles of the 54 candidate genes were validated using RT-PCR, initially on RNA isolated from a range of normal human tissues obtained post mortem, including testicular RNA. Of the 54 genes, 21 were expressed in more than two non-testis/non-CNS normal tissues and were therefore dismissed at this stage. Of the remaining 33 genes (bold in Supplementary Table), 30 had expression limited to the testis in normal tissue, 2 had expression limited to the testis and normal CNS tissues and 1 further gene had expression in one or two normal tissues in addition to testis, with or without CNS expression.
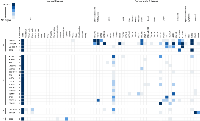
The same 33 genes, which showed predominant (or only) expression in the testis (bold in Supplementary Table), were then analysed by RT-PCR in a range of cancer cell types. Following this analysis, 14 of these genes were shown to have no expression in any of the cancerous material (Supplementary Table). The expression profiles for those genes exhibiting cancer cell expression are shown in Figure 1. A further 16 genes were shown to have expression in at least one cancerous tissue and no expression in normal cells other than testicular cells (Figure 1, class B). Of the remaining 3 genes, 2 were cancer/testis/CNS restricted [i.e. expressed in at least one cancer cell type, in addition to the testis and normal CNS tissue (Figure 1, class C)] and 1 was cancer/testis-selective [i.e. expressed in one or two normal tissues other than CNS, as well as the testis and at least one cancer type (Figure 1, class D)].
Meta-analysis of candidate genes expression profiles
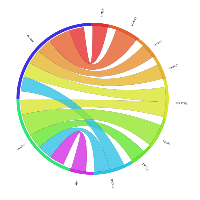
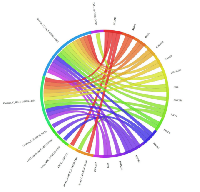
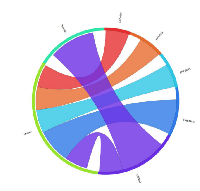
In order to explore the possible clinical relevance of the newly identified genes, we conducted meta-analyses using patient-derived cancer microarray data, including 13 cancer types in a range of 80 microarray data sets [48]. Firstly, we investigated the expression profiles of 18 of the 19 genes that exhibited expression in at least one of the cancer cell types tested by RT-PCR (1 was not present on the microarrays -TGIF2LX). Of these, 9 showed meta-up-regulation in either ovarian and/or prostate cancers (50.0%; Figure 2). An example of the meta-up-regulation of an individual gene (SPZ1) is given in the Forest plot profile for ovarian cancer in Figure 3. Whilst the meta-analysis reveals 9 genes to be up-regulated for two given cancer types (ovarian, prostate), analysis of single cancer data sets from the 80 cancer data sets used reveals evidence for activation of a total of 15 of the 18 candidate genes in at least one patient-derived sample set (83.3%; Figure 4).
Of the 33 genes with meiCT gene potential (based on expression patterns in normal tissue), 14 genes did not appear to be expressed in any of the cancer cell types analysed by RT-PCR (Supplementary Table). To further explore the possibility that these genes are meiCT genes, we used 11 of the 14 genes for meta-analyses using the 80 cancer gene expression microarray data sets (3 were not present on the microarrays, C1orf141, HEATR7B and SATL1) and found expression profiles for 5 of these genes were indicative of a cancer type marker for ovarian and prostate cancers (45.5%; Figure 5). A further 5 (10 genes in total) were expressed in at least one single cancer data set (Figure 6), indicating the potential to mark a specific sub-group of tumours within a cancer type. Only C8orf74 exhibited no measurable expression in cancer cells / tissues (although C1orf141, HEATR7B and SATL1 could not be analysed via meta-analyses due to their absence on the arrays).
DISCUSSION
CTAs are cancer-specific biomarkers with considerable potential in prognostics/diagnostics and as therapeutic targets. The current classification system for CTA genes continues to be based on that put forward by Hoffman and colleagues [40]. We have since proposed a sub-category of CTA genes based on an in silico pipeline originating with putative meiotic genes; we have termed these meiCT genes [44]. Here we describe a further 29 genes which are novel meiCT genes. As for previously characterised meiCT genes, we found that most of these new genes are autosomally encoded (28 out of 29; Supplementary Table), a finding consistent with the transcriptional inactivation of the X chromosome during male meiosis [45, 46]. An additional commonality with the previously characterised meiCT genes was the fact that many of this new cohort were shown to be up-regulated as a general marker for ovarian cancers: 10 of the new cohort displaying a meta-change increase in gene expression were in ovarian cancer. This again raises the possibility of using the meiCT genes to improve the diagnosis of this diverse and pernicious cancer type. It may be the case that genes that have a normal biological function (i.e. meiotic) in the foetal ovary are preferentially reactivated in cancers of this tissue type. Additionally, ovarian cancers are currently most frequently treated with cytoreductive surgery and chemotherapy although these types of tumours are immunoreactive and there is currently extensive work ongoing to explore the application of immune-based therapies for their treatment [49]. Thus, the identification of ovarian cancer-specific markers such as these is of exceptional potential therapeutic value [49].
Recent work has demonstrated that sub-groups of 26 germline and placental specific genes can be used to delineate aggressive metastasis-prone lung cancers [30]. This indicates that small sub-groups of tissue-specific genes can serve as accurate biomarkers in the stratification of complex and heterogeneous cancers. The clinical implications of this are far reaching as they offer extensive potential in establishing best practice approaches to therapeutic stratification. This work provides a paradigm for how germline gene expression in cancers can be applied to clinical stratification of complex disease. Having a definitive on/off expression profile, as observed with the meiCT genes, greatly enhances the potential simplicity of application of these genes in novel prognostics technologies.
A number of studies have now specifically explored the potential of expression of human germline genes as cancer biomarkers. Interestingly, whilst common genes have been identified however, the various studies have all identified additional distinct genes indicating that the full mining of data sets of this magnitude require multiple and diverse approaches. For example, this current study has identified 32 new genes with tight germline and germline/CNS tissue-specific expression restrictions (of the 33 genes analysed, BOLL was unique in exhibiting expression in two other somatic tissues, so was classed as testis selective); however, a recent seminal and extensive study of human male germline/placental genes only identified 21 of the 32 (65.6%) genes reported here as germline -specific [30]. A slightly lower trend is seen when analysing previously reported meiCT genes (22 out of 52 meiCT genes (42.3%) were reported as germline genes [30]; a total of 52.4% for all reported meiCT genes [44 and data presented here]).
In addition to serving as cancer biomarkers the meiCT genes may serve as therapeutic targets via a variety of routes. Firstly, the immunogenicity of the gene products of the meiCT genes remains very poorly characterised. Their highly stringent tissue specificity infers that their gene products could potentially serve as tumour-specific immunotherapeutic targets. Given the heterogeneity of cancer, both intra- and inter-tumour, the development of a large bank of markers/immunotherapeutic targets will be of increasing importance in personalisation strategies [5, 50-52].
Lastly, germline genes in D. melanogaster serve to drive the oncogenic programme [38]. The human orthologues of these genes are also widely activated in human tumours [36] and other CTA genes have been demonstrated to be required for cancer cell proliferation and their depletion can serve to sensitise cancer cells to standard therapeutic agents (for example, see 33,34). Thus, not only can germline gene products potentially offer direct targets for drug therapies, but depletion of their activity can also serve to enhance the efficacy of existing therapies potentially enabling reduced dose regimes, which will limit undesired drug toxicities. The cancer-specific nature of meiCT gene expression makes these genes exceptionally attractive for further exploration in drug targeting and drug sensitisation.
The suggestion that germline genes are oncogenic infers that some of the genes identified here could play a tumour initiating / progression role. One of the genes validated here is a member of the ADAM gene family - ADAM2. The ADAM proteins often exhibit proteolytic activity and have emerging roles in the invasive properties of specialist cells within the placenta [53]. Such proteolytic functions may promote invasion and metastasis of solid tumours. A putative role for other members of the adamalysins in the aetiology and pathology of colorectal cancer and melanoma has been proposed [54, 55]. This might indicate that not only are germline genes required for oncogenesis, they might drive metastasis and/or invasion and thus offer cancer-specific intervention points to stop the lethal spread of tumours. SPZ1 was shown to be testis-specific in our normal tissue panel, a finding previously shown by Hsu and colleagues [56]. In their study, they further showed that the gene was expressed both in the testis and epididymis. We found positive expression in a colon and ovarian cancer cell line and on meta-analysis there was a significant up-regulation in ovarian cancer. It has since been shown by Hsu and colleagues that SPZ1, which encodes a transcription factor, acts as a proto-oncogene to promote cellular proliferation and tumour formation in a mouse model [57]. Despite this, SPZ1 is not a previously recognised CTA gene [18].
It has been suggested that another of the novel meiCT genes identified here, SHCBP1L, encodes a protein with strong homology to a mouse protein present in proliferating cells and may have similar physiological effects [58]. It remains unexplored whether this protein indeed acts through similar signal transduction pathways to promote proliferation but as the gene has shown a statistically significant meta-change up-regulation, again in ovarian cancer, makes this possibility worthy of further exploration.
Closing remarks
Germline genes are emerging as important cancer-specific factors. The extent of their clinical importance is only now starting to come into focus, either as biomarkers for stratification and diagnosis, as oncogenic activators and as drug or immunotherapeutic targets. What is becoming increasingly apparent is the heterogeneity of tumour cell populations. This is the driver for the need to identify a large cohort of cancer cell markers that individually or in combination can target and mark a large number of tumour types and cell populations. Here we identify a new and extensive cohort of genes that can contribute to the growing catalogue of bona fide cancer-specific biomarkers.
MATERIALS AND METHODS
Cell lines and cell culture
The following cell lines were purchased from the European Collection of Cell Cultures (ECACC); 1321N1, COLO800, COLO857, G-361, HCT116, HT29, LoVo, MM127, SW480 and T84. The H460 cell line was purchased from the American Type Culture Collection (ATCC), and the two ovarian adenocarcinoma cell lines, PEO14 and TO14, were obtained from Cancer Research Techology Ltd. The NTERA-2 (clone D1) cell line was gifted by Prof. P.W. Andrews (University of Sheffield) and is regularly authenticated within the group using standard antibody tests using anti-OCT4 antibodies and retinoic acid-induced differentiation. The A2780 cell line was provided by Prof. P. Workman (Cancer Research UK Centre for Cancer Therapeutics, Surrey, UK) and was authenticated at source. Primary cultures of proliferating human prostate smooth muscle cells were obtained from PromoCellTM(C-12574). All cultures were used within a six months period of obtaining validated lines from external sources.
1321N1, A2780, NTERA-2 (clone D1) and SW480 cell lines were cultured in Dulbecco’s modified Eagle’s medium from InvitrogenTM (DMEM + GLATAMAXTM) supplemented with 10% foetal bovine serum (FBS). COLO800, COLO857 and H460 cell lines were cultured in Invitrogens Roswell Park Memorial Institute 1640 medium (RPMI 1640) + GLUTAMAXTM with 10% FBS. PEO14 and TO14 cell lines were cultured in RPMI 1640 + GLUTAMAXTM supplemented with 10% FBS and 2mM sodium pyruvate, and MM127 was cultured in RPMI 1640 + GLUTAMAXTM supplemented with 10% FBS and 25mM HEPES. McCoy’s 5A medium + GLUTAMAXTM supplemented with 10% FBS was used to culture the G-361, HCT116 and HT29 cell lines. Ham’s F12 + DMEM (1:1) + GLUTAMAXTM (InvitrogenTM) with 10% FBS was used to culture T84 cells.
All cell lines were grown in a 37°C incubator with 5% CO2, with the exception of the NTERA-2 (clone D1) cell line, which was grown at 37°C with 10% CO2.
cDNA construction
Total RNA preparations were obtained from the human normal tissue panels (ClontechTM; 636643). RNA from tumour tissues and cell lines were purchased from ClontechTM and AmbionTM. Total RNA was also isolated from cells using TRIzol (Invitrogen). Confluent cells were collected in TRIzol reagent and incubated at room temperature for 5 minutes. Chloroform was added with vigorous shaking and incubated for 5 minutes at room temperature. The aqueous phase was transferred to a clean tube following centrifugation at 12,000 g for 15 minutes at 4°C. The RNA was precipitated out of solution using isopropanol (10 minutes at room temperature and centrifuged at 12,000 g for 20 minutes at 4°C). RNA preparations were re-suspended in RNase-free water containing DNase. The concentration and quality of RNA was measured using a NanoDrop (ND_1000). 1.0 µg of total RNA was reverse-transcribed into cDNA using the SuperScript III First Strand synthesis kit (InvitrogenTM) as per the manufacturer’s instructions.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
The sequences for each of the genes analysed were obtained from the National Center for Biotechnology (NCBI; http://www.ncbi.nlm.nih.gov/). Forward and reverse primers for each of the genes were designed, and where possible were intron-spanning.
A volume of 2 µl of diluted cDNA (containing ~150 ng/µl cDNA) was used for PCR in a 50 µl final volume. BioMixTM Red and MyTaqTM Red (BiolineTM) was used for PCR amplification. Samples were amplified with a pre-cycling hold at 96°C for 5 minutes, followed by 40 cycles of denaturing at 96°C for 30 seconds, annealing at a temperature between 54 and 60°C for 30 seconds and extensions at 72°C for 40 seconds followed by a final extension step at 72°C for 5 minutes. The products were separated on 1% agarose gels containing ethidium bromide or Gel GreenTM.
ACKNOWLEDGEMENTS
SJS was supported by a Wales Clinical Academic Training Fellowship from the Welsh Assembly Government and North West Cancer Research (project grant CR950). JF was supported by the National Institute of Social Care and Health Research (grant HS/09/008). RJM and JAW were funded by Cancer Research Wales. RJM was funded by North West Cancer Research (project grants CR888 and CR950).
Conflict of Interest
There are no conflicts of interest to declare.
- 1. Bailey AM, Mao Y, Zeng J, Holla V, Johnson A, Brusco L, Chen K, Mendelsohn J, Routbort MJ, Mills GB, Meric-Bernstam F. Implementation of biomarker-driven cancer therapy: existing tools and remaining gaps. Discov Med. 2014; 17: 101-114. [PubMed].
- 2. Clinical proteomic biomarkers: relevant issues on study design & technical considerations in biomarker development. Clin Trans Med. 2014; 3: 7. https://doi.org/10.1186/2001-1326-3-7. [PubMed].
- 3. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol. 2014; in press. https://doi.org/10.3109/09553002.2014.892229. [PubMed].
- 4. Stratified medicine in European Medicines Agency licensing: a systematic review of predictive biomarkers. BMJ Open. 2014; 4: e004188. https://doi.org/10.1136/bmjopen-2013-004188. [PubMed].
- 5. Personalizing oncology: perspectives and prospects. J Clin Oncol. 2013; 31: 1904-1911. https://doi.org/10.1200/JCO.2012.45.3605. [PubMed].
- 6. The long journey of cancer biomarkers from the bench to the clinic. Clin Chem. 2013; 59: 147-157. https://doi.org/10.1373/clinchem.2012.184614. [PubMed].
- 7. Serologic autoantibodies as diagnostic cancer biomarkers-a review. Cancer Epidemiol Biomarkers Prev. 2013; 22: 2161-2181. https://doi.org/10.1158/1055-9965.EPI-13-0621. [PubMed].
- 8. The perspective of immunotherapy: new molecules and new mechanisms of action in immune modulation. Curr Opin Oncol. 2014; 26: 204-214. https://doi.org/10.1097/CCO.0000000000000054. [PubMed].
- 9. Reprogramming antitumour immunity. Trends Immunol. 2014; S1471-4906: 00028-3. https://doi.org/10.1016/j.it.2014.02.003. [PubMed].
- 10. Manipulating immune cells for adoptive immunotherapy of cancer. Curr Opin Immunol. 2014; 27C: 46-52. https://doi.org/10.1016/j.coi.2014.01.008. [PubMed].
- 11. Cancer immunotherapy – revisited. Nat Rev Drug Discov. 2011; 10: 591-600. https://doi.org/10.1038/nrd3500. [PubMed].
- 12. Cancer immunotherapy comes of age. Nature. 2011; 480: 480-489. https://doi.org/10.1038/nature10673. [PubMed].
- 13. In search of magic bullets: the golden age of immunotherapeutics. Cell. 2014; 157: 227-240. https://doi.org/10.1016/j.cell.2014.03.010. [PubMed].
- 14. Adoptive immunotherapy for cancer. Immunol Rev. 2014; 257: 14-38. https://doi.org/10.1111/imr.12136. [PubMed].
- 15. Advances in cancer immunotherapy. Discov Med. 2013; 15: 120-125. [PubMed].
- 16. A personalized view on cancer immunotherapy. Cancer Lett. 2013; S0304-3835: 00670-8. https://doi.org/10.1016/j.canlet.2013.09.016. [PubMed].
- 17. Antibody-based immunotherapy of cancer. Cell. 2012; 148: 1081-1084. https://doi.org/10.1016/j.cell.2012.02.034. [PubMed].
- 18. CTdatabase: a knowledgebase of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009; 37: D816-819. https://doi.org/10.1093/nar/gkn673. [PubMed].
- 19. Caballero OL, Chen YT. Cancer/testis antigens: potential targets for immunotherapy. Cancer Sci. 2009; 100: 2014-2021. https://doi.org/10.1111/j.1349-7006.2009.01303.x. [PubMed].
- 20. Cancer/testis (CT) antigens, carcinogenesis and spermatogenesis. Spermatogenesis. 2011; 1: 209-220. https://doi.org/10.4161/spmg.1.3.17990. [PubMed].
- 21. Concise review: cancer/testis antigens, stem cells, and cancer. Stem Cells. 2007; 25: 707-711. https://doi.org/10.1634/stemcells.2006-0469. [PubMed].
- 22. The biology of cancer testis antigens: putative function, regulation and therapeutic potential. Mol Oncol. 2011; 5: 164-182. https://doi.org/10.1016/j.molonc.2011.02.001. [PubMed].
- 23. Cancer-testis antigens: the current status on antigen regulation and potential clinical use. Am J Blood Res. 2012; 2: 29-35. [PubMed].
- 24. Cancer testis antigens: novel biomarkers and targetable proteins for ovarian cancer. Int Rev Immunol. 2011; 30: 127-137. https://doi.org/10.3109/08830185.2011.572504. [PubMed].
- 25. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005; 5: 615-625. https://doi.org/10.1038/nrc1669. [PubMed].
- 26. The cancer/testis genes: Review, standardization, and commentary. Cancer Immun. 2004; 4: 1. [PubMed].
- 27. Causes and consequences of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol. 2014; 54: 251-272. https://doi.org/10.1146/annurev-pharmtox-011112-140326. [PubMed].
- 28. The testis in immune privilege. Immunol Rev. 2006; 213: 66-81. https://doi.org/10.1111/j.1600-065X.2006.00438.x. [PubMed].
- 29. Tight junctions in the testis: new perspectives. Philos Trans R Soc Lond B Biol Sci. 2010; 365: 1621-1635. https://doi.org/10.1098/rstb.2010.0010. [PubMed].
- 30. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone ling cancers. Sci Transl Med. 2013; 5: 186ra66. https://doi.org/10.1126/scitranslmed.3005723. [PubMed].
- 31. The role of cancer-testis antigens as predictive and prognostic markers in non-small cell lung cancer. PLoS One. 2013; 8: e67876. https://doi.org/10.1371/journal.pone.0067876. [PubMed].
- 32. Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, Yee C. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med. 2008; 358: 2698-2703. https://doi.org/10.1056/NEJMoa0800251. [PubMed].
- 33. Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Mol Cell Biol. 2012; 32: 4131-4140. https://doi.org/10.1128/MCB.00686-12. [PubMed].
- 34. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007; 446: 815-819. https://doi.org/10.1038/nature05697. [PubMed].
- 35. Induced malignant genome reprogramming in somatic cells by testis-specific factors. Biochim Biophys Acta. 2011; 1809: 221-225. https://doi.org/10.1016/j.bbagrm.2011.04.003. [PubMed].
- 36. Feichtinger J, Larcombe L, McFarlane RJ. Meta-analysis of expression of l(3)mbt tumor-associated germline genes supports the model that a soma-to-germline transition is a hallmark of human cancers. Int J Cancer. 2014; 134: 2359-2365. https://doi.org/10.1002/ijc.28577. [PubMed].
- 37. Sustaining cancer through addictive ectopic gene activation. Curr Opin Oncol. 2014; 26: 73-77. https://doi.org/10.1097/CCO.0000000000000032. [PubMed].
- 38. Ectopic activation of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010; 330: 1824-1827. https://doi.org/10.1126/science.1195481. [PubMed].
- 39. Potential role of meiosis proteins in melanoma chromosomal instability. J Skin Cancer. 2013; 2013: 190109. https://doi.org/10.1155/2013/190109. [PubMed].
- 40. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci USA. 2008; 105: 20422-20427. https://doi.org/10.1073/pnas.0810777105. [PubMed].
- 41. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001; 61: 5544-5551. [PubMed].
- 42. Shiohama Y, Ohtake J, Ohkuri T, Noguchi D, Togashi Y, Kitamura H, Nishimura T. Identification of a meiosis-specific protein, MEIOB, as a novel cancer/testis antigen and its augmented expression in demethylated cancer cells. Immunol Lett. 2014; 158: 175-182. https://doi.org/10.1016/j.imlet.2014.01.004. [PubMed].
- 43. Identification of a meiosis-specific protein as a member of the class of cancer/testis antigens. Proc Natl Acad Sci USA. 1998; 95: 5211-5216. https://doi.org/10.1073/pnas.95.9.5211. [PubMed].
- 44. Meta-analysis of clinical data using human meiotic genes identifies a novel cohort of highly restricted cancer-specific marker genes. Oncotarget. 2012; 3: 843-853. https://doi.org/10.18632/oncotarget.580. [PubMed].
- 45. Meiotic sex chromosome inactivation. Curr Biol. 2010; 20: R962-963. https://doi.org/10.1016/j.cub.2010.09.041. [PubMed].
- 46. Meiotic sex chromosome inactivation. Development. 2007; 134: 1823-1831. https://doi.org/10.1242/dev.000018. [PubMed].
- 47. CancerEST: a web-based tool for automatic meta-analysis of public EST data. Database. 2014; 2014: bau024. https://doi.org/10.1093/database/bau024. [PubMed].
- 48. CancerMA: a web-based tool for automatic meta-analysis of public cancer microarray data. Database. 2012; 2012: bas055. https://doi.org/10.1093/database/bas055. [PubMed].
- 49. Tse BWC, Collins A, Oehler MK, Zippelius A, Heinzelmann-Schwarz VA. Antibody-based immunotherapy for ovarian cancer: where are we at? Ann Oncol. 2014; 25: 322-331. https://doi.org/10.1093/annonc/mdt405. [PubMed].
- 50. Seoane J, De Mattos-Arruda L. The challenge of intratumour heterogeneity in precision medicine. J Intern Med. 2014. https://doi.org/10.1111/joim.12240. [PubMed].
- 51. Swanton C. Cancer evolution: the final frontier of precision medicine? Ann Oncol. 2014; 25: 549-551. https://doi.org/10.1093/annonc/mdu005. [PubMed].
- 52. Individualized medicine from prewomb to tomb. Cell. 2014; 157: 241-253. https://doi.org/10.1016/j.cell.2014.02.012. [PubMed].
- 53. Pollheimer J, Fock V, Knöfler M. Review: The ADAM metalloproteinases – novel regulators of trophoblast invasion? Placenta. 2014; 35S: S57-S63. https://doi.org/10.1016/j.placenta.2013.10.012. [PubMed].
- 54. ADAM and ADAMTS family proteins and their role in the colorectal cancer etiopathogenesis. BMB Rep. 2013; 46: 139-150. https://doi.org/10.5483/BMBRep.2013.46.3.176. [PubMed].
- 55. Metalloproteinases in melanoma. Eur J Cell Biol. 2014; S0171-9335: 00003-X.
- 56. Spz1, a novel bHLH-Zip protein, is specifically expression in testis. Mech Dev. 2001; 100: 177-187. https://doi.org/10.1016/s0925-4773(00)00513-x. [PubMed].
- 57. Hsu SH, Hsieh-Li HM, Huang HY, Huang PH, Li H. bHLH-zip transcription factor Spz1 mediates mitogen-activated protein kinase cell proliferation, transformation, and tumorigenesis. Cancer Res. 2005; 65: 4041-4050. https://doi.org/10.1158/0008-5472.CAN-04-3658. [PubMed].
- 58. Cloning and characterisation of 13 novel transcripts and the human RGS8 gene from the 1q25 region encompassing the hereditary prostate cancer (HPC1) locus. Genomics. 2001; 73: 211-222. https://doi.org/10.1006/geno.2001.6500. [PubMed].
Last Modified: 2016-06-06 14:17:05 EDT
PII: 37
