Oncoscience
Abstract | PDF | Full Text | Supplementary Material | How to Cite
https://doi.org/10.18632/oncoscience.333
BRG1 and BRM loss selectively impacts RB and P53, respectively: BRG1 and BRM have differential functions in vivo
Stefanie B. Marquez-Vilendrer1, Sudhir K. Rai1, Sarah JB Gramling1, Li Lu1,2, David N. Reisman1
1 Department of Hematology/Oncology, Medicine, University of Florida, Gainesville, FL, USA
2 Department of Pathology, University of Florida, Gainesville, FL, USA
Correspondence to: David N. Reisman, email:[email protected]
Keywords: retinoblastoma, lung cancer, p53, swi/snf, tumor suppressor
Received: August 29, 2016
Accepted: September 23, 2016
Published: December 21, 2016
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
The SWI/SNF complex is an important regulator of gene expression that functions by interacting with a diverse array of cellular proteins. The catalytic subunits of SWI/SNF, BRG1 and BRM, are frequently lost alone or concomitantly in a range of different cancer types. This loss abrogates SWI/SNF complex function as well as the functions of proteins that are required for SWI/SNF function, such as RB1 and TP53. Yet while both proteins are known to be dependent on SWI/SNF, we found that BRG1, but not BRM, is functionally linked to RB1, such that loss of BRG1 can directly or indirectly inactivate the RB1 pathway. This newly discovered dependence of RB1 on BRG1 is important because it explains why BRG1 loss can blunt the growth-inhibitory effect of tyrosine kinase inhibitors (TKIs). We also observed that selection for Trp53 mutations occurred in Brm-positive tumors but did not occur in Brm-negative tumors. Hence, these data indicate that, during cancer development, Trp53 is functionally dependent on Brm but not Brg1. Our findings show for the first time the key differences in Brm- and Brg1-specific SWI/SNF complexes and help explain why concomitant loss of Brg1 and Brm frequently occurs in cancer, as well as how their loss impacts cancer development.
INTRODUCTION
The SWI/SNF complex is an essential regulator of a large cadre of genes. By moving and shifting the position of histones within the chromatin, SWI/SNF gives key cellular proteins and transcription factors access to specific DNA domains necessary to regulate gene expression [1]. This complex is composed of two mutually exclusive catalytic subunits, either Brahma (BRM or SMARCA2) or Brahma Related Gene 1 (BRG1 or SMARCA4), along with 8-10 subunits that assemble into at least three different but related SWI/SNF complexes [2, 3]. SWI/SNF subunits with similar functionality, such ARID1A versus ARID1B or BAF60A, B, and C versus BAF53 A and B, in combination with BRG1 or BRM, give SWI/SNF both the flexibility and diversity characteristic of the many different molecular complexes that can be assembled. Knowledge of the interplay of these subunits with cellular proteins is ever-expanding; however, it is not fully understood how the loss of SWI/SNF subunits truly impacts cancer development through the additional loss of expression of the other subunits, which are frequently altered in cancer.
Through its various protein interactions, the SWI/SNF complex has been linked to many cellular processes, including growth control, differentiation, development, adhesion, and DNA repair [4, 5]. It is not surprising, then, that this complex and its subunits are targeted during cancer development. Recent Next Generation (NextGen) sequencing studies show that at least one SWI/SNF subunit is mutated in 20% of all human cancers [6]. Certain subunits are highly mutated in specific cancers such as the following: PBRM1 (BAF180) is mutated in liver and renal carcinoma [7, 8], while ARID1A is preferentially mutated in uterine, cervical, ovarian and gastric cancers [9-11]. The function of many of these subunits, however, is only partially understood in contrast to the function of BRG1 and BRM. These subunits serve as the ATPase catalytic or mechanical motor of the SWI/SNF complex, and the loss of one or both clearly abrogates the ability of SWI/SNF to open the chromatin and change gene expression [4, 5]. This, together with the fact BRG1 and BRM are more broadly silenced in a range of cancers compared with other subunits, signifies that the study of the BRG1 and BRM subunits is important to the field of cancer research.
SWI/SNF, BRG1, and BRM have been linked to the function of a number of key cellular proteins required to thwart cancer development. In particular, SWI/SNF is known to be essential for RB1-mediated growth inhibition. Specifically, cell lines that lack both BRG1 and BRM are refractory to growth inhibition when a constitutively active form of RB1 is introduced [12, 13]. RB1-mediated growth inhibition can be restored if either BRG1 or BRM is also restored along with this introduction of RB1 [14, 15]. Moreover in BRG1/BRM-deficient cell lines, the re-expression of BRG1 or BRM readily stimulates growth inhibition [16-18], yet high levels of BRG1 or BRM used in in vitro experiments can activate both BRG1-dependent and BRM-dependent SWI/SNF complexes, and thus it is not clear whether RB1 is dependent on BRG1 or BRM complexes. This represents a major gap in our knowledge. Methods to restore BRG1 or BRM expression could be pursued as a novel avenue of targeted therapy, and therefore the determination of whether either BRG1- or BRM-dependent complexes or both are functionally tied to RB1 is critical. This is also potentially clinically important, as tyrosine kinase inhibitors (TKIs) arrest growth in part through the activation of RB1 [19], and knowledge of whether BRM, BRG1, or both is functionally tied to RB1 allows one to determine if the loss of either or both of these subunits might cause resistance to TKIs.
Similar to RB1, TP53 has been linked to the function of a number of subunits of SWI/SNF, including BRG1 and BRM [20, 21]. If BRG1 or BRM or both can substitute for RB1 and TP53, one might predict an inverse relationship of the mutation rates of TP53 and RB1 with those of BRG1 and BRM, as determined by NextGen sequencing studies. However, unlike TP53, which is principally inactivated by mutations, BRM is almost never mutated (<2%) but, rather, it is epigenetically silenced [22]. Similarly, the frequency of BRG1 mutations is also relatively low (<2-5%) in most tumors [22], and our recent work has shown that BRG1 can be silenced by either aberrant splicing or translational blocking mechanisms regulated by the AKT pathway [23]. Interestingly, ARID1A, which is usually inactivated by mutation, does indeed show an inverse correlation with TP53 mutations in a number of tumor types [24-26]. Thus, like RB1, it is unclear if BRG1 or BRM-dependent complexes are functionally linked to TP53, or if both are linked to this protein. The oncogene-induced senescence hypothesis [27, 28] was designed to explain how cells develop into benign tumors and why they evolve into malignant tumors if and when both RB1 and TP53 proteins are abrogated. Since we know that SWI/SNF is functionally linked to both RB1 and TRP53, we predicted that tumors would more readily arise if BRG1 and BRM are concomitantly lost. Specifically, we wanted to determine if Brg1, Brm or both substitute for Rb1 and Trp53 in a murine model of lung cancer.
METHODS
Generation of murine lung adenocarcinoma
Quad mice (see tandem manuscript for definition of genotypes), which carried the CCSP-rtTA and (oTet)7-Cre constructs (generous gifts from Jeff Whittsett [29, 30]) were used; these constructs drive the targeted inactivation of a floxed gene. Quad mice were homozygous for the Brm-null (Brm-/-) allele and for floxed-Brg1 (Brg1fLoxP) alleles (gifts from Moshe Yaniv and Pierre Chambon, respectively) [31, 32]. We also generated Cre-negative mice, which were similar to Quad mice, but lacked the (oTet)7-Cre construct and the Brm-null alleles. When the Quad mice were crossed with mice with Cre-negative phenotypes, an “Intermediate” strain resulted, in which mice were homozygous for the LoxP-Brg1 gene and the CCSP-rtTA construct, but were hemizygous for the (oTet)7-Cre constructs and the Brm-null allele. After these intermediate mice were crossed with each other and after the heterozygous Brm-null offspring were removed from further study, we generated four genotypes as follows: wild type (WT: Brm+/Brg1+), Brm-null (Brm-/Brg1+), Brg1-negative (Brg1-/Brm+), and double-negative (Brg1-/Brm-). At 6-7 weeks of age, all mice were given 2 intra-peritoneal (IP) injections (1 week apart) of 1 g/kg urethane (ethyl carbamate) to initiate tumor development. These injections also served to prevent apoptosis caused by Cre-mediated Brg1 inactivation in normal lung cells (i.e., in type 2 alveolar and Clara cells) [33]. Four weeks after the first IP injection, the mice were provided ad libitum with water containing 1 mg/mL tetracycline and 3% sucrose for 5 days to induce Cre expression and thus inactivate the Brg1 allele. At 6-12 months, the mice were euthanized due to the development physical stress, at which point lung adenocarcinomas were harvested and analyzed.
Immunohistochemistry and Immunofluorescence
Hematoxylin & eosin (H&E) staining was performed to assess the general histology of the tumors and for scoring purposes. Antibodies to the following antigens were used in IHC/Immunofluorescence (IF) experiments: Brg1 (sc-374197, 1:50, Santa Cruz Biotechnology, Dallas, TX, USA); Brg1 (21634-1-AP, 1:250, Protein Tech, Chicago, IL, USA); Brm (1:200; rabbit polyclonal antibody generated by the Reisman Lab); Rb1 (ab6075, 1:100, Abcam, Cambridge, MA, USA); pRb1S780 (9307S, 1:200, Cell Signaling Technology, Danvers, MA, USA); pRb1T821/826 (sc-16669, 1:50, Santa Cruz Biotechnology, Dallas, TX, USA); Pcna (610664, 1:300, BD Biosciences, San Jose, CA, USA); Pcna (RB-9055-P0, 1:200, Thermo Scientific, Waltham, MA, USA); Ki-67 (550609, 1:100, BD Biosciences); cyclin D1 (sc-753, 1:50, Santa Cruz Biotechnology); Trp53 (sc-6243, 1:50, Santa Cruz Biotechnology); anti-Cdk2, GTX22363 (1:200, GeneTex); Cdk4 (12790, 1:100, Cell Signaling Technology). All antibodies were tested for cross-reactivity (specificity) by staining cell lines that lack the antigen of interest; these tests were then confirmed by western blot. All tissue sections were subjected to antigen retrieval, which consisted of heating in a microwave for 15 minutes on the high setting using either 10 mM sodium citrate buffer (pH 6), 10 mM Tris buffer (pH 10) or 10 mM Tris, 1 mM EDTA and 0.05% Tween20 (pH 8), depending on the antibody. Slides were incubated either overnight at 4°C or for 2 hours at room temperature. The appropriate biotinylated secondary antibodies were then used at a 1:200 dilution (BA-1000 or BA-9200, Vector Labs, Burlingame, CA, USA). This was followed by incubation with horseradish peroxidase streptavidin for 1 hour at room temperature (SA-5004, 1:200, Vector Labs). DAB was used as the chromogen (550880, BD Pharmingen, San Jose, CA, USA), and Harris hematoxylin was used as the counterstain. For IF experiments, the secondary antibodies Alexa-fluor 488 and Alexa-fluor 594 (A1100 and A21207, respectively, Life Technologies, Carlsbad, CA, USA) were used at dilutions of 1:200.
Western Blot
Our western blot method has been described elsewhere [34, 35]. Briefly, the cells were harvested, and total protein was extracted using a urea-based lysis buffer, as described previously. Proteins were subjected to gel electrophoresis and then transferred onto polyvinylidene difluoride membranes. The membranes were incubated overnight at 4°C or at room temperature for 1 hour with the following primary antibodies: anti-Brg1, 21634-1-AP (1:1000, Protein Tech), phospho-Gsk3β (Ser9) (Cell Signaling Technology #9336), anti-Rb, sc-73598 (1:200, Santa Cruz Biotechnology), anti-p21, 2947P (1:1000, Cell Signaling Technology), anti-cyclin D1, 2978P (1:1000, Cell Signaling Technology), anti-Cdk2, GTX22363 (1:200, GeneTex) and anti-Gapdh, GTX100118 (1:1000, GeneTex). The pRb antibodies used in the western blot are the same as those described in the above section. The membranes were then incubated for 1 hour at room temperature with the appropriate biotinylated anti-mouse or anti-rabbit secondary antibody, which were used at a dilution of 1:2000 (GE Healthcare Life Sciences, Pittsburgh, PA, USA). Proteins were detected with a Chemiluminescence Western Bright ECL kit (K-12045-D50, Advansta Inc., Menlo Park, CA, USA).
Sequencing
PFU enzyme and Trp53 primers were used to amplify cDNA from the total RNA extracted from the tumors. Similarly, individual cDNA clones were amplified with Pfu polymerase, A-tailed and sequenced using M13 forward and reverse primers following cloning into a pGEM T Easy vector. (Note: Pfu produces blunt ends so that the PCR product must be A-tailed for cloning into T vector). Bands were isolated, gel-purified (QIAQuick Gel Extraction Kit, Qiagen, Valencia, CA, USA) and submitted for Sanger sequencing at the sequencing core of the Interdisciplinary Center for Biotechnology Research at the University of Florida.
RESULTS
Trp53 mutations are not detected in tumors derived from Brm-null mice.
While activated oncogenes drive the initial development of lung adenomas, senescence of these premalignant growths is known to be induced by Rb1 and Trp53, which block their further transformation into adenocarcinomas [36]. As such, the transformation from adenoma to adenocarcinoma occurs via the abrogation of the senescence effects of Trp53 and Rb1 [27, 28, 37]. However, if Brg1 and Brm are indeed necessary cofactors for the function of Rb1 and Trp53, we would expect that this transformation would also be fostered by the inactivation Brg1, Brm or both. Previous studies have established a dependency of the apoptotic and cell cycle arrest functions of Trp53 upon interaction with and specific binding to SWI/SNF subunits [20, 38-40]. However, it is unclear if Trp53 dependence on SWI/SNF stems from its interaction with Brg1- or with Brm-dependent complexes. As mutations in Trp53 are commonly found in murine lung adenocarcinomas, we investigated whether there was a change in the observed Trp53 mutation rate in these lung adenocarcinomas as a function of Brm or Brg1 loss.
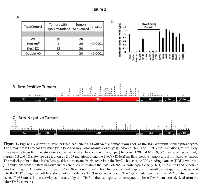
In our murine lung cancer model, we observed malignant adenocarcinomas in all four genotypes used in this experimental system, with a frequency of 99%, 80%, 73% and 48% for the DKO, Brg1-KO, Brm-null and wild type genotypes, respectively (as described in our tandem paper). To determine possible functional relationships between Brg1 and Brm proteins with Trp53, we determined the rate of Trp53 mutations in each of the four genotypes by sequencing the total mRNA from these tumors. The sequencing of Trp53 (n=20 for each genotype) revealed that 80.0% (16/20) and 66.6% (13/20) of tumors from WT and Brg1-KO genotypes (Brm-positive tumors), respectively, harbored Trp53 missense mutations (Figure 1A); differences in the mutation rates between these two groups were not statistically significant (p>0.05). In comparison, Trp53 mutations occurred in only 5% (1/20) and 0% (0/20) of tumors from Brm-null and DKO mice (Brm-negative tumors), respectively (Figure 1A). The difference between the Trp53 mutation rates in tumors from the Brm-positive and Brm-negative phenotypes was statistically significant (p<0.01). Of the mutations found in the WT (~69) and Brg1-KO (~63) tumor cells, we determined that 56/69 (~80%) and 47/63 (~75%) Trp53 mutations occurred between amino acids 100-300, which is the DNA binding domain (or the TRP53 mutation hot spot in humans). The observed lack of Trp53 mutations in the Brm-negative phenotype was not likely due to loss of Trp53 expression via p19Arf1 mutations or Mdm2 amplifications, since Trp53 was expressed in the vast of majority of tumor cells (>85%) by IHC and was not qualitatively different between Brm-negative tumors and Brm-positive tumors (Supplementary Figure 1). Interestingly, the lack of observable Trp53 mutations has also been shown in ovarian and gastric tumors that also demonstrate the loss of another SWI/SNF subunit, ARID1A [24-26].Brm loss blocks selection of Trp53 mutations
The above data do not preclude the possibility that Trp53 mutations develop within tumor cells, but rather, the data suggest that such mutations might occur in less than 5% of tumor cells and thus are not detectible via Sanger sequencing. To test this hypothesis, we sequenced individual Trp53 cDNA clones (70 each) from WT- and Brm-null-derived tumors. We observed similar frequencies of non-synonymous (missense) Trp53 mutations of 79% and 58% from the WT and Brm-null phenotypes, respectively (p>0.05). These mutations that were detected in individual cDNA clones from tumors derived from the Brm-null phenotypes appeared to be randomly distributed throughout the Trp53 open reading frame (Figure 1C). In contrast, the Trp53 mutations that were detected by Sanger sequencing (cDNA clones and total mRNA) from wild type Brm-positive tumors appeared to be clustered in a known Tp53 mutational hotspot DNA binding domain (DBD) (Figure 1B). These data suggest that Trp53 mutations arose in individual tumor cells in mice of both Brm-positive and Brm-negative phenotypes, but that in the Brm-positive tumor cells, the clones of more dysfunctional Trp53 mutations became more ubiquitous (positive selection) within a given Brm-positive tumor. As the percentage of cells with these mutations increased within the tumors (>5-10% of the tumor), this in turn allowed the mutations to become detectible by standard Sanger sequencing.
We sought to determine whether Trp53 mutations within the DBD in general lacked function regardless of whether they were derived from either Brm genotype; similarly, we sought to determine whether Trp53 mutations from the other domains retained TRP53 function. We tested 14 clones identified from both Brm-negative and Brm-positive tumors. Of these clones, we tested 7 from the DNA binding domain and 7 from outside this region (Supplementary Figure 2). As expected, the Trp53 mutant cDNA clones from the DBD showed little to no transcriptional function in a Trp53-dependent luciferase reporter assay, regardless of the genotype of origin. In comparison, the cDNA clones from outside the DBD largely retained Trp53 function (Figure 1D and Supplementary Figure 2). Therefore, the more tumorigenic mutations from the DBD only arose by selection if they occurred in Brm-positive tumors. In contrast, Trp53 mutations that arose from all regions in Brm-negative tumors, regardless if they caused an inactivation of Trp53 function, failed to undergo selection and did not result in an increase in the percentage of tumor cells expressing a given Trp53 mutation. Based on these data, loss of Brm expression causes loss of Trp53 selection/evolution during tumor development.
Loss of Brg1 substitutes for Rb1 inactivation, but Brm loss does not affect Rb1 phosphorylation.
We next examined whether loss of Brg1, Brm, or both affected how the Rb1 pathway became inactivated. In vitro data have shown that BRG1 and BRM are necessary cofactors for RB1 function, where RB1 poorly inhibits growth in the absence of a functional SWI/SNF complex, and where the re-expression of either BRG1 or BRM in BRG1/BRM-deficient cell lines restores RB1-mediated growth inhibition [14, 15]. Although these in vitro experiments demonstrate the functional dependency of RB1 on either BRG1 or BRM, it is not clear if RB1 specifically depends on BRG1- or BRM-specific complexes in vivo.
Using these lung tumor phenotypes, we sought to determine whether Rb1 is functionally linked to Brg1, Brm or both by examining the Rb1 phosphorylation status as a function of Brg1/Brm expression. To accomplish this, we performed dual immunofluorescence (IF) staining using an anti-Brg1 or anti-Brm antibody with two different anti-phosphoRb1 (pRb1) antibodies to reflect Rb1 activity or inactivity. Specifically, hyperphosphorylation at Rb1-Ser780 (pRb1S780) occurs by mitogenic stimulation, while hypophosphorylation (pRb1hypo) occurs during G1 arrest [41]. Phosphorylation at Rb1-Thr821/826 (pRB1T821/826) causes a conformational change, which prevents the binding of Rb1 to proteins that contain the LXCXE sequence [42]. Therefore, these two phospho-Rb1 antibodies give an indirect measurement of Rb1 functionality or lack thereof.Brm loss does not correlate with Rb1 phosphorylation
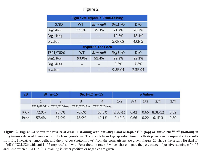
We first conducted IHC for total Rb1 expression to exclude the loss of Rb1, and we observed that Rb1-negative tumor cells accounted for less than 5% of all tumor cells in tumors from each of the four genotypes, which is consistent with previously published reports [43]. Next, dual IF staining was conducted with anti-Brm and anti-pRb1S780 antibodies to determine the phosphorylation state of Rb1 as a function of Brm expression. We found that the frequencies of Rb1 hyperphosphorylation in Brm-positive (WT mice) and Brm-negative tumors (Brm-null mice) were similar (~75%; p>0.05) (Figure 2A, top). Similarly, using antibodies to Brm and pRb1T821/826, we found no significant difference in pRb1T821/826 staining (~90% p>0.5) as a function of Brm expression (Figure 2A, bottom). Hence, Brm loss did not affect or change the phosphorylation status of Rb1. Brg1 loss is linked to the hypophosphorylation of Rb1
We then examined the phosphorylation status of Rb1 in Brg1-positive tumor cells using dual IF with antibodies against Brg1 and pRb1S780 in tumors from each of the four genotypes. Although Brg1-KO tumors were not necessarily completely devoid of Brg1 expression, the majority of tumor cells from these genotypes were Brg1-negative (variable mosaic pattern). By dual IF, we observed the frequency of pRb1S780 staining to be significantly and statistically lower in Brg1-negative tumor cells derived from Brg1-KO mice compared with Brg1-positive cells from WT tumors (19.3% vs. 77.1%: p=5E-8) (Figure 2A). In fact, Brg1-positive cells in tumors from each of the four genotypes showed significantly more pRb1S780 immunoreactivity than Brg1-negative tumor cells from Brg1-KO mice (p<0.0001). In a parallel experiment, dual IF with antibodies against Brg1 and pRb1T821/826 was conducted. With this pRb1 antibody, we again obtained statistically significant results showing that Rb1 was more likely to be hypophosphorylated (i.e., in its active state) in Brg1-negative cells (Brg1-KO) compared with Brg1-positive (WT) cells (Figure 2A) (9.5% vs. 92.3%: p=9.8E-4). Importantly, IHC confirmed the infrequent loss of total Rb1 in these tumors (<5% of tumor cells in a given tumor were negative for Rb1) (data not shown). As Rb1 is not frequently lost in these tumors, these data demonstrate that Brg1 loss correlates with the hypophosphorylated state of Rb1. Is Brg1 loss sufficient to disrupt the Rb1 pathway?
The loss of p16 (CDKN2A) and the overexpression of cyclin D/E and/or Cdk2/4 drive inactivation of the Rb1 pathway by causing Rb1 to become hyperphosphorylated (pRB1Hyper). Brg1-negative tumor cells primarily harbored hypophosphorylated Rb1 (pRb1Hypo), so none of the above mechanisms of Rb1 inactivation (pRb1Hyper) accounts for how the Rb1 pathway is inactivated in Brg1-negative tumor cells. Additionally, the infrequent loss of total Rb1 in these tumors eliminates that as a major cause for disruption of the Rb1 pathway. Previous work by multiple investigators has shown that Rb1-mediated growth inhibition requires a functional Brg1 protein [12-15]. As such, the in vivo loss of Brg1 might be sufficient to inactivate the Rb1 pathway. As shown in Figure 2A, we observed that only ~19% and ~9% of Brg1-negative tumor cells were also positive for pRb1S780 and/or pRb1T821/826 , respectively, by dual IF; in other words, ~80% and ~90% of Brg1-negative cells failed to express either pRb1S780 or pRb1T821/826 and also harbor hypophosphorylated or activated Rb1, which should inhibit growth. We hypothesized that if Brg1 loss does indeed inactivate the Rb1 pathway, Brg1-negative cells should continue to proliferate even though they harbor active Rb1. To test this hypothesis, we needed to determine definitively whether these pRb1Hypo-positive/Brg1-negative tumor cells were proliferating and were not growth-arrested.Is Pcna/Ki67 expression similar in pRb1Hypo-expressing Brg1-KO tumor cells and in pRb1Hyper-expressing WT tumor cells?
In order to determine if Brg1-negative/pRb1Hypo tumor cells were proliferating or if they were growth-arrested, we conducted dual IF to detect two different markers of proliferation (Ki67 or Pcna) together with the anti-pRb1T821/826 antibody. We compared the percentage of Pcna immunoreactivity in WT tumor cells that expressed pRb1Hyper (inactive Rb1) and those that did not (i.e., expressed pRb1hypo) (Figure 2B). We observed that Pcna staining was significantly higher at 57.4% in WT pRb1Hyper-Brg1 positive tumor cells versus 34% (p=3.4E-3) in pRb1Hypo-Brg1-positive WT tumor cells. This indicates that in WT tumor cells, when Rb1 is hypophosphorylated, tumor cells grow more slowly. In comparison, in Brg1-KO tumor cells, we found no difference in Pcna immunoreactivity between pRb1T821/826-positive and pRb1T821/826-negative tumor cells (45.9% vs. 49.1%, respectively, p=0.66). We observed a similar pattern with antibodies against Ki67 and anti-pRb1T821/826 (Figure 2B). A statistically significant decrease in Ki67 immunoreactivity was observed in the WT Brg1-positive tumor cells that did not express pRb1T821/826 compared with WT Brg1-positive tumor cells that did express pRb1T821/826 (positive) (26% and 72.0%, respectively; p=2.0E-11). Again, this finding indicates that WT tumor cells that express pRbHypo grow more slowly when Brg1 is present. In comparison, in the Brg1-KO tumor cells, we did not observe a statistically significant difference in Ki67 staining between pRb1T821/826-negative and pRb1T821/826-positive tumor cells (70.6% vs. 68.0%, respectively, p=0.42). Our results indicate that proliferation of WT Brg1-positive tumor cells is slowed in the presence of pRb1Hypo while Brg1-deficient tumors grow equally well in the presence of either pbB1Hypo or pRb1Hyper. BRG1 loss regulates the phosphorylation of RB1
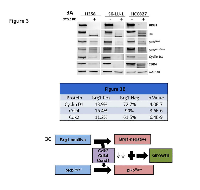
The connection between BRG1 loss and RB1 phosphorylation suggests a relationship between these two proteins. Hence, we examined the impact of BRG1 knockdown via an shRNA approach in 3 lung cancer-derived cell lines (H358, SK-LU-1 and HCC-827) to determine the potential effects on RB1 phosphorylation. By western blot, BRG1 knockdown caused an observed shift from upper phosphorylated bands to lower non-phosphorylated bands, which indicates a change from an inactive RB1 (pRB1Hyper) state to an active one (pRB1Hypo), as well as a decrease in phosphorylation at RB1 sites T821/826 and S780. BRG1 knockdown also caused the downregulation of CCND1 (cyclin D1), CDK2 and CDK4 (Figure 3A), which has been previously reported [44-47]. The expression of CCNE1, CDKN1A (p21) and CDKN2A (p16) either did not appear to change or did not change in a similar direction in all 3 lung cancer cell lines as a function of BRG1 expression (data not shown).
Next, we analyzed the expression of these cell cycle-associated genes by qPCR in tumors derived from WT and Brg1-KO mice. While we observed little to no difference in the expression levels of p16 (Cdkn2a) and Ccne1 in tumors of these genotypes, we did observe a decrease in Cdk4 (4.8-fold p<0.01) and a decrease in Ccnd1 mRNA (~5.8-fold; p<0.01) in the Brg1-KO tumor cells compared with the WT tumor cells. To confirm that changes in Ccnd1 and Cdk4 contribute to the decrease in Rb1 phosphorylation in murine Brg1-deficient lung tumors, we conducted dual IF for Brg1 versus both Ccnd1 and Cdk4. We observed that Ccnd1 was expressed in 13.9% of Brg1-negative tumor cells and was expressed in 72.7% of Brg1-positive tumor cells; this difference was statistically significant (p=4.0E-7) (Figure 3B). Similarly, Cdk4 was expressed in 16.4% of Brg1-negative tumor cells and in 90% of Brg1-positive tumor cells; this difference was also statistically significant (p=3.6E-6) (Figure 3B). In addition, by dual IF, we observed a statistical correlation between Cdk2 loss and Brg1 loss (11.2% and 61.5% in Brg1-KO and WT tumors, respectively; p=6.4E-9). These data indicate that Brg1 loss is linked to pRb1Hypo in part through decreases in Cdk2, Cdk4 and Ccnd1 expression (Figure 3C). In addition, the observed decrease in Rb1 phosphorylation in these murine adenocarcinoma cells was not associated with a decrease in the expression of the proliferation markers Pcna and Ki67, which indicates that the Rb1 pathway is likely unable to inhibit growth. These data therefore suggest that Brg1 loss indirectly or directly blocks the function of the Rb1 pathway, which allows cancer cells that express pRb1Hypo to continue to proliferate. This is consistent with in vitro data that show that Brg1 is a required cofactor for Rb1-mediated growth inhibition in human tumor-derived cell lines.
Brg1 loss blocks tyrosine kinase inhibitor-induced RB1-mediated growth inhibition
To date, the RB1 pathway in human cancers has been reported to be disrupted only by loss of expression of CDKN2A or RB1, as well as by CDK4/CCND1 over-expression [48], even though SWI/SNF is known to be an essential cofactor for RB1 function [16, 17]. This is due in part because it has never been clear whether BRG1 or BRM is essential for RB1, although both have been shown to be functionally linked to RB1 [14, 15]. Based on the understanding that RB1 is essentially only connected with BRG1, we could now pursue the impact of BRG1 loss on RB1-dependent processes. To this end, most TKIs inhibit growth in part by activating RB1-mediated growth inhibition [49, 50]. One would therefore predict that loss of SWI/SNF complex activity, and more specifically the loss of BRG1 expression, would abrogate the ability of RB1 to inhibit growth and thereby block the general ability of TKIs to halt tumor growth. To test this prediction, we tested two TKIs, Sorafenib (a RAF-1, B-RAF and VEGFR-2 inhibitor) and Picropodophyllin (PPP, an inhibitor of insulin-like growth factor receptor: IGF-1R). We treated two BRG1-deficient cancer cell lines that harbored either scrambled or anti-BRG1 shRNA with either Sorafenib or PPP for 96 hours and measured the Figure 3.
Figure 3A shows the protein expression levels as measured by western blot with antibodies to BRG1, RB1, pRB1S780, pRB1T821/826, CCND1, CDK4 and GAPDH for the three human cell lines H358, SK-LU-1 and HCC827 with (+) and without (-) BRG1 knockdown. Also shown is the effect of BRG1 suppression by shRNA knock down on RB1 phosphorylation. Cell proliferation persists (before or after BRG1 knock down) since either RB1 or BRG1 is inactivated and therefore cannot cooperate to foster RB1-mediated growth inhibition in either situation. Figure 3B shows the results of dual IF for Brg1 and Ccnd1 in the top row and the results for dual IF between Brg1 and Cdk4 and Brg1 and Cdk2 in the middle and bottom rows, respectively; p values for these comparisons are given in the column on the far right. Figure 3C illustrates the changes in Cdk2, Cdk4 and Ccnd1 as a function of Brg1 loss and how the loss of Brg1 potentially allows the Brg1-negative tumor cells to continue to grow, despite being in its active growth-inhibitory state.
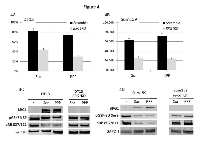
expression of BRG1 and the number of cells over time to determine the growth rate. We observed that both compounds readily induced BRG1 as well as reduced GSK3β phosphorylation (Figure 4C and 4D), and inhibited the growth of Sum159 and DT13, two BRG1-deficient breast cancer cell lines (Figure 4A and 4B). However, if these cell lines harbored anti-BRG1 shRNA, we observed that BRG1 was not induced and GSK3β phosphorylation was virtually unchanged, as measured by western blot (Figure 4C and 4D). This lack of change in GSK3β phosphorylation is important because the hypo-phosphorylated GSK3β is the active form, which fosters CCND1 degradation and causes p21 to translocate to the nucleus [51, 52]. Both of these changes cause RB1 to become hypophosphorylated and active, thereby inhibiting growth. Hence, the lack of BRG1 expression blocks RB1-mediated growth inhibition by two different possible mechanisms. First, the lack of BRG1 could directly or indirectly block GSK3β dephosphorylation, which in turns inhibits the dephosphorylation and activation of RB1. This can be seen in Figure 4C and 4D row 3, where pRB1807/811 decreases with BRG1 induction but remains relatively the same when BRG1 induction is blocked. Second, BRG1 is known to be an essential cofactor for RB1/E2F-mediated gene expression [53], and without BRG1, RB1/E2F cannot effectively induce gene expression of target genes required to block G1 to S phase transition. It is not surprising then that we observed that these compounds were not nearly as effective in inducing growth inhibition in these cell lines when BRG1 expression was suppressed as compared with when BRG1 was readily induced. Moreover, we show that the loss of BRG1 expression can cause resistance to TKI-mediated growth inhibition in vitro and thus may contribute to the clinically observed resistance to these and other TKIs in vivo.
DISCUSSION
In vitro experimental data from a number of different labs have clearly linked both TP53 and RB1 function to the SWI/SNF complex [4, 5]. However, whether TP53 and RB1 are functionally linked to BRG1- or BRM-dependent SWI/SNF complexes has been a subject of debate. Moreover, in vitro overexpression experiments are problematic in addressing this question because the strong viral promoters often used yield such high levels that the ectopic expression of BRG1 or BRM activates both BRG1- and BRM-dependent SWI/SNF complexes [15]. Further, shRNA knockdown experiments do not completely deplete BRG1 and BRM in BRG1/BRM-positive cell lines, and low levels of BRG1/BRM can still have an impact on cell phenotypes [23]. It has therefore been difficult to differentiate the separate functions of BRG1 versus BRM. For this reason, the in vivo knockout of Brg1 and Brm is better positioned to address the functional differences between Brg1 and Brm. To this end, our data clearly show that Brg1 loss impacts the Rb pathway directly or indirectly and allows tumor cells that harbor hypophosphorylated Rb1 to proliferate as fast as or faster than tumor cells harboring hyperphosphorylated Rb1. RB1 and BRG1 are further linked, as BRG1 loss downregulates CDKs and G1 cyclins to cause a change in RB1 phosphorylation. This change does not cause redistribution of cells in the cycle cell due to growth arrest, as these tumor cells continue to proliferate. The way in which BRG1 regulates cyclins/CDKs is not known, but data from our lab have shown that BRG1 is linked to GSK3β (published data), which regulates the cellular location of p21 and the stability of CCND1 [52, 54, 55].
The relationship between SWI/SNF and RB1 was first demonstrated by experiments performed by Dunaief and Strober [16, 17]. In these experiments, the re-expression of BRG1 induced growth inhibition, which was dependent on RB1, and to a lesser degree, on the RB1 family members p107 and p130. Subsequently, both BRG1 and BRM were shown to possess LXCXE domains that bind RB1 directly [56]. Several labs have shown that BRG1 re-expression can restore RB1 function in BRG1/BRM-deficient cell lines [12, 13]. We and others have shown that restoring BRM expression is equally effective at the restoration of RB1 function [14, 15]. Hence, in vitro data have shown that BRG1 and BRM play similar and potentially overlapping roles in the mediation of RB1 function. However, our data strikingly demonstrate that BRG1 loss, but not BRM loss, appears to directly or indirectly contribute to the conversion of RB1 into its hypophosphorylated state. Therefore, BRG1 loss potentially blocks RB1-mediated growth inhibition in vivo. Why is establishing that BRG1 is linked to RB1 important? BRG1 loss is not yet recognized as a major mechanism of RB1 pathway inactivation, in part because BRG1 loss occurs concomitantly with tumor cell loss of CDKN2A (p16) or RB1. Hence, the interdependence of BRG1 on RB1 might be viewed as unimportant to cancer. However, there are a number of drugs used clinically, such as TKIs and CDK inhibitors that inhibit growth in part by activating RB1 [50, 57]. In our previous work, we showed that BRG1 could be silenced by aberrant splicing (irreversibly silenced) or translationally blocked (reversibly silenced) [23], as in the Sum159 and DT13 cell lines. Hence, the treatment of these cancer cells, where BRG1 is reversibly silenced, with TKIs (e.g., Sorafenib and PPP) appears to induce BRG1 and inhibit growth. However, when BRG1 induction is blocked by anti-BRG1 shRNA knockdown, the cells demonstrate resistance to growth inhibition, which occurs when BRG1 can be induced. It is important to recognize that knockdown by shRNA cannot completely suppress BRG1 expression, and therefore, the loss of growth inhibition may be greater when BRG1 expression is completely abrogated by mutation or aberrant BRG1 splicing. Given Figure 4. this interdependence of BRG1 on RB1, it should not be surprising that BRG1 loss can impart resistance to TKIs (as illustrated by our data). Further research will be needed to determine which TKIs are affected by BRG1 loss.
In contrast to the potential resistance caused by BRG1 inactivation, a number of different investigators have reported increased sensitivity to taxanes and DNA-damaging agents such as cisplatin and carboplatin [58, 59]. Moreover, as BRG1 is required for double-strand repair, its loss may also enhance the sensitivity of tumors to PARP inhibitors. In support of this assertion is the observation that inactivation of the SWI/SNF subunit ARID1A indeed causes increased sensitivity to PARP inhibitors. Furthermore, BRG1 has been linked to the function of BRCA1, and if true, this could further explain how BRG1 loss might enhance sensitivity to PARP inhibitors. These data help us understand why the connection between BRG1 and RB1 as well as the link between BRG1 and DNA repair have potential clinical consequences in determining sensitivity or resistance to specific drugs used clinically. Hence, BRG1 expression or lack thereof may be an important biomarker to determine which drugs should be clinically used.
Similar to RB1, TP53 functionality has also been linked to SWI/SNF [21], and like RB1, it is unclear if TP53 is functionally linked to BRG1, BRM or both proteins in vivo. We showed that Trp53 mutations occur in our murine model system, which is consistent with other carcinogen-induced murine lung tumor models [60-62]. By Sanger sequencing, we found tumor-driving Trp53 mutations in both wild type and Brg1-deficient tumors, but Trp53 mutations were not found in Brm-negative tumors. However, the absence of Trp53 mutations by sequencing total mRNA does not mean that they were not present, but rather, it indicates that they were below the threshold of detection by Sanger sequencing. Our data demonstrate that Trp53 mutations are indeed generated by the chemical carcinogen used in this model system in both Brm-positive and Brm-negative tumor cells. When Brm was present, these chemically generated Trp53 mutations appeared to undergo selection pressure during tumor development, whereas those Trp53 mutations (typically in the DNA binding domain of Trp53) that lack functionality (Trp53-mediated transcription) were positively selected. This idea is consistent with the oncogene-induced senescence hypothesis, where malignant tumors only arise in those tumor cells where Trp53 is inactivated. However, we found that, in contrast, malignant tumors arose in Brm-deficient tumors regardless of the type of Trp53 mutation present in the tumor cells. Our findings indicate that Brm loss appears to usurp the driving force that selects for non-functional Trp53 isoforms. Thus, one can deduce that Brm-dependent complexes are linked to Trp53. As such, one might predict that Trp53 mutations would not occur concomitantly with Brm mutations. However, Brm is not commonly silenced by mutation. It is important, therefore, to remember that Brm is part of a complex, and while Brm itself may not be mutated, other subunits might show a relationship with Trp53 mutations. This appears to be the case in human ovarian clear cell carcinomas and endometrioid endometrial cancers, where the loss of ARID1A appears to be nearly mutually exclusive from TP53 mutations [24, 26].
The functional relationship between TP53 and BRM may be important as it may explain how BRM polymorphisms are predictive of cancer risk [22], and thus foster early steps of transformation. BRM silencing is known to be driven by two insertional 6-base-pair polymorphisms contained within the BRM promoter [63]. These polymorphic sites serve as functional binding sites for a number of proteins (HDAC3, HDAC9 and MEF2D) that underlie the epigenetic silencing of BRM [64]. As a loss of TP53 function via nonsense or missense mutation is known to be transforming, cancer initiation may occur when BRM polymorphisms drive BRM silencing, which in turn fosters transformation by thwarting or impairing TP53 function. In tumors or cell lines, BRM silencing is not known to occur concomitantly with TP53 mutations. The clinical relevance of the link between BRM complexes to TP53 has not been fully delineated. Nevertheless, the connection between BRM complexes and TP53 gives us a deeper understanding of how SWI/SNF is involved in cancer initiation and progression and it generates new questions as to how SWI/SNF is involved in cancer.
CONFLICTS OF INTEREST
No conflicts of interest were disclosed.
- 1. Muchardt C and Yaniv M. When the SWI/SNF complex remodels...the cell cycle. Oncogene. 2001; 20(24):3067-3075. https://doi.org/10.1038/sj.onc.1204331. [PubMed].
- 2. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr Top Microbiol Immunol. 2003; 274:143-169. https://doi.org/10.1007/978-3-642-55747-7_6. [PubMed].
- 3. SWI/SNF chromatin remodeling complexes and cancer. American journal of medical genetics Part C, Seminars in medical genetics. 2014; 166C(3):350-366. https://doi.org/10.1002/ajmg.c.31410. [PubMed].
- 4. Alteration to the SWI/SNF complex in human cancers. Oncology Reviews. 2010; 4(2).
- 5. The SWI/SNF complex and cancer. Oncogene. 2009; 28(14):1653-1668. https://doi.org/10.1038/onc.2009.4. [PubMed].
- 6. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013; 45(6):592-601. https://doi.org/10.1038/ng.2628. [PubMed].
- 7. PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer J. 2013; 19(4):324-332. https://doi.org/10.1097/PPO.0b013e3182a102d1. [PubMed].
- 8. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat Commun. 2015; 6:6120. https://doi.org/10.1038/ncomms7120. [PubMed].
- 9. The roles of ARID1A in gynecologic cancer. Journal of gynecologic oncology. 2013; 24(4):376-381. https://doi.org/10.3802/jgo.2013.24.4.376. [PubMed].
- 10. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol Rep. 2016; 35(2):607-613. https://doi.org/10.3892/or.2015.4421. [PubMed].
- 11. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012; 33(1):100-103. https://doi.org/10.1002/humu.21633. [PubMed].
- 12. BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci U S A. 2000; 97(14):7748-7753. https://doi.org/10.1073/pnas.97.14.7748. [PubMed].
- 13. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000; 101(1):79-89. https://doi.org/10.1016/S0092-8674(00)80625-X. [PubMed].
- 14. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem. 2002; 277(7):4782-4789. https://doi.org/10.1074/jbc.M109532200. [PubMed].
- 15. Concomitant downregulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002; 21(8):1196-1207. https://doi.org/10.1038/sj.onc.1205188. [PubMed].
- 16. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996; 16(4):1576-1583. https://doi.org/10.1128/mcb.16.4.1576. [PubMed].
- 17. Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR and Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994; 79(1):119-130. https://doi.org/10.1016/0092-8674(94)90405-7. [PubMed].
- 18. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000; 60(21):6171-6177. [PubMed].
- 19. RB1 dual role in proliferation and apoptosis: cell fate control and implications for cancer therapy. Oncotarget. 2015; 6(20):17873-17890. https://doi.org/10.18632/oncotarget.4286. [PubMed].
- 20. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem. 2007; 282(52):37429-37435. https://doi.org/10.1074/jbc.M706039200. [PubMed].
- 21. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002; 277(25):22330-22337. https://doi.org/10.1074/jbc.M111987200. [PubMed].
- 22. Beyond Mutations: Additional Mechanisms and Implications of SWI/SNF Complex Inactivation. Front Oncol. 2014; 4:372. https://doi.org/10.3389/fonc.2014.00372. [PubMed].
- 23. Marquez-Vilendrer1 S, Thompson K and Reisman D. Mechanism of BRG1 Silencing in Primary Cancers. Oncotarget. 2016 Jul 13. https://doi.org/10.18632/oncotarget.10593. [PubMed].
- 24. Bosse T, ter Haar NT, Seeber LM, v Diest PJ, Hes FJ, Vasen HF, Nout RA, Creutzberg CL, Morreau H and Smit VT. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod Pathol. 2013; 26(11):1525-1535. https://doi.org/10.1038/modpathol.2013.96. [PubMed].
- 25. Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, Lee SP, Ho SL, Chan AK, Cheng GH, Roberts PC, Rejto PA, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011; 43(12):1219-1223. https://doi.org/10.1038/ng.982. [PubMed].
- 26. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011; 71(21):6718-6727. https://doi.org/10.1158/0008-5472.CAN-11-1562. [PubMed].
- 27. Oncogene- and tumor suppressor gene-mediated suppression of cellular senescence. Semin Cancer Biol. 2011; 21(6):367-376. https://doi.org/10.1016/j.semcancer.2011.10.005. [PubMed].
- 28. Oncogene-induced cellular senescence. Adv Anat Pathol. 2010; 17(1):42-48. https://doi.org/10.1097/PAP.0b013e3181c66f4e. [PubMed].
- 29. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic research. 2002; 11(1):21-29. https://doi.org/10.1023/a:1013986627504. [PubMed].
- 30. Conditional recombination reveals distinct subsets of epithelial cells in trachea, bronchi, and alveoli. American journal of respiratory cell and molecular biology. 2005; 33(5):455-462. https://doi.org/10.1165/rcmb.2005-0180OC. [PubMed].
- 31. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997; 17(10):5976-5986. https://doi.org/10.1128/mcb.17.10.5976. [PubMed].
- 32. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). Embo J. 1998; 17(23):6979-6991. https://doi.org/10.1093/emboj/17.23.6979. [PubMed].
- 33. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008; 68(10):3689-3696. https://doi.org/10.1158/0008-5472.CAN-07-6652. [PubMed].
- 34. Identifying targets for the restoration and reactivation of BRM. Oncogene. 2013; 33:653-664. https://doi.org/10.1038/onc.2012.613. [PubMed].
- 35. Flavonoids from Each of the Six Structural Groups Reactivate BRM, A Possible Co-factor for the Anti-Cancer Effects of Flavonoids. Carcinogenesis. 2014; 35(10):2183-2193. https://doi.org/10.1093/carcin/bgu117. [PubMed].
- 36. MYC synergizes with activated BRAFV600E in mouse lung tumor development by suppressing senescence. Cancer Res. 2014; 74(16):4222-4229. https://doi.org/10.1158/0008-5472.CAN-13-3234. [PubMed].
- 37. Malkinson AM. Primary lung tumors in mice: an experimentally manipulable model of human adenocarcinoma. Cancer Res. 1992; 52(9 Suppl):2670s-2676s. [PubMed].
- 38. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene. 2009; 28(27):2492-2501. https://doi.org/10.1038/onc.2009.121. [PubMed].
- 39. BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem. 2008; 283(18):11924-11934. https://doi.org/10.1074/jbc.M705401200. [PubMed].
- 40. BAF53 interacts with p53 and functions in p53-mediated p21-gene transcription. J Biochem. 2007; 142(5):613-620. https://doi.org/10.1093/jb/mvm176. [PubMed].
- 41. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997; 17(10):5771-5783. https://doi.org/10.1128/mcb.17.10.5771. [PubMed].
- 42. Molecular mechanisms underlying RB protein function. Nature reviews Molecular cell biology. 2013; 14(5):297-306. https://doi.org/10.1038/nrm3567. [PubMed].
- 43. Molecular aspects of chemical carcinogenesis: the roles of oncogenes and tumour suppressor genes. Toxicology. 1995; 96(3):173-194. https://doi.org/10.1016/0300-483x(94)02991-3. [PubMed].
- 44. BRG1 expression is increased in human cutaneous melanoma. Br J Dermatol. 2010; 163(3):502-510. https://doi.org/10.1111/j.1365-2133.2010.09851.x. [PubMed].
- 45. Regulation of PTEN expression by the SWI/SNF chromatin-remodelling protein BRG1 in human colorectal carcinoma cells. Br J Cancer. 2011; 104(1):146-154. https://doi.org/10.1038/sj.bjc.6606018. [PubMed].
- 46. BRG1 expression is increased in human glioma and controls glioma cell proliferation, migration and invasion in vitro. J Cancer Res Clin Oncol. 2012. https://doi.org/10.1007/s00432-012-1172-8. [PubMed].
- 47. BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS One. 2013; 8(3):e59772. https://doi.org/10.1371/journal.pone.0059772. [PubMed].
- 48. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene. 2002; 21(45):6908-6914. https://doi.org/10.1038/sj.onc.1205834. [PubMed].
- 49. and Ramos KS. The promise and failures of epigenetic therapies for cancer treatment. Cancer treatment reviews. 2014; 40(1):153-169. https://doi.org/10.1074/jbc.M705401200. [PubMed].
- 50. Reversible G(1) arrest induced by inhibition of the epidermal growth factor receptor tyrosine kinase requires up-regulation of p27(KIP1) independent of MAPK activity. The Journal of biological chemistry. 2000; 275(10):6987-6995. https://doi.org/10.1074/jbc.275.10.6987. [PubMed].
- 51. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell cycle. 2003; 2(4):339-345. [PubMed].
- 52. GSK-3beta regulates cyclin D1 expression: a new target for chemotherapy. Cellular signalling. 2008; 20(4):581-589. https://doi.org/10.1016/j.cellsig.2007.10.018. [PubMed].
- 53. Singh S, Johnson J and Chellappan S. Small molecule regulators of Rb-E2F pathway as modulators of transcription. Biochim Biophys Acta. 2010; 1799(10-12):788-794. https://doi.org/10.1016/j.bbagrm.2010.07.004. [PubMed].
- 54. Phosphatidylinositol 3’-kinase, mTOR, and glycogen synthase kinase-3beta mediated regulation of p21 in human urothelial carcinoma cells. BMC urology. 2011; 11:19. https://doi.org/10.1186/1471-2490-11-19. [PubMed].
- 55. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes & development. 1998; 12(22):3499-3511. https://doi.org/10.1101/gad.12.22.3499. [PubMed].
- 56. Role of the LXCXE binding site in Rb function. Mol Cell Biol. 2000; 20(18):6799-6805. https://doi.org/10.1128/mcb.20.18.6799-6805.2000. [PubMed].
- 57. Ling YH, Li T, Yuan Z, Haigentz M, Jr., Weber TK and Perez-Soler R. Erlotinib, an effective epidermal growth factor receptor tyrosine kinase inhibitor, induces p27KIP1 up-regulation and nuclear translocation in association with cell growth inhibition and G1/S phase arrest in human non-small-cell lung cancer cell lines. Molecular pharmacology. 2007; 72(2):248-258. https://doi.org/10.1124/mol.107.034827. [PubMed].
- 58. SMARCA4/BRG1 is a novel prognostic biomarker predictive of cisplatin-based chemotherapy outcomes in resected non-small cell lung cancer. Clin Cancer Res. 2015. https://doi.org/10.1158/1078-0432.CCR-15-1468. [PubMed].
- 59. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp Cell Res. 2012; 318(16):1973-1986. https://doi.org/10.1016/j.yexcr.2012.06.011. [PubMed].
- 60. Horio Y, Chen A, Rice P, Roth JA, Malkinson AM and Schrump DS. Ki-ras and p53 mutations are early and late events, respectively, in urethane-induced pulmonary carcinogenesis in A/J mice. Mol Carcinog. 1996; 17(4):217-223. https://doi.org/10.1002/(SICI)1098-2744(199612)17:4<217::AID-MC5>3.0.CO;2-A. [PubMed].
- 61. Characterization of p53 mutations in methylene chloride-induced lung tumors from B6C3F1 mice. Carcinogenesis. 1993; 14(5):803-810. https://doi.org/10.1093/carcin/14.5.803. [PubMed].
- 62. Rb and p53 gene deletions in lung adenocarcinomas from irradiated and control mice. Radiat Res. 1997; 148(1):81-89. [PubMed].
- 63. Two novel BRM insertion promoter sequence variants are associated with loss of BRM expression and lung cancer risk. Oncogene. 2011; 30(29):3295-3304. https://doi.org/10.1038/onc.2011.81. [PubMed].
- 64. Kahali B, Yu J, Marquez SB, Thompson KW, Liang SY, Lu L and Reisman D. The silencing of the SWI/SNF subunit and anticancer gene BRM in Rhabdoid tumors. Oncotarget. 2014; 5(10):3316-3332. https://doi.org/10.18632/oncotarget.1945. [PubMed].
Last Modified: 2017-01-05 11:41:14 EST
