Oncoscience
ATRX, IDH1-R132H and Ki-67 immunohistochemistry as a classification scheme for astrocytic tumors
Jinquan Cai1,6,*, Chuanbao Zhang2,3,*, Wei Zhang2,3, Guangzhi Wang1,6, Kun Yao6,7, Zhiliang Wang3,6, Guanzhang Li2,6, Zenghui Qian2,6 Yongli Li1,6, Tao Jiang2,3,4,5,6, Chuanlu Jiang1,6
1Department of Neurosurgery, The Second Affiliated Hospital of Harbin Medical University, NanGang District, Harbin,
Heilongjiang Province 150001, China
2Beijing Neurosurgical Institute,Capital Medical University,Dongcheng District,Beijing 100050,
China
3Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Dongcheng District, Beijing 100050, China
4Beijing Institute for Brain Disorders Brain Tumor Center,Dongcheng District,Beijing 100050,China
5China National Clinical Research Center for Neurological Diseases, Dongcheng District, Beijing100050, China
6Chinese Glioma Cooperative Group(CGCG), Dongcheng District, Beijing 100050, China
7Department of Pathology, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing100093, China
*These authors have contributed equally to the work
Correspondence to: Yongli Li, email: [email protected]
Keywords: ATRX, IDH-R132H, Ki-67, astrocytic tumors, progression
Received: May 10, 2016
Accepted: August 12, 2016
Published:September 06, 2016
Chuanlu Jiang, email: [email protected] Tao Jiang, email: [email protected]
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Recurrence and progression to higher grade lesions are key biological events and characteristic behaviors in the evolution process of glioma. Malignant astrocytic tumors such as glioblastoma (GBM) are the most lethal intracranial tumors. However, the clinical practicability and significance of molecular parameters for the diagnostic and prognostic prediction of astrocytic tumors is still limited. In this study, we detected ATRX, IDH1-R132H and Ki-67 by immunohistochemistry and observed the association of IDH1-R132H with ATRX and Ki-67 expression. There was a strong association between ATRX loss and IDH1-R132H (p<0.0001). However, Ki-67 high expression restricted in the tumors with IDH1-R132H negative (p=0.0129). Patients with IDH1-R132H positive or ATRX loss astrocytic tumors had a longer progressive-free survival (p<0.0001, p=0.0044, respectively). High Ki-67 expression was associated with shorter PFS in patients with astrocytic tumors (p=0.002). Then we characterized three prognostic subgroups of astrocytic tumors (referred to as A1, A2 and A3). The new model demonstrated a remarkable separation of the progression interval in the three molecular subgroups and the distribution of patients’ age in the A1-A2-A3 model was also significant different. This model will aid predicting the overall survival and progressive time of astrocytic tumors’ patients.
INTRODUCTION
Astrocytic tumors are the most common group of human gliomas with inherent tendency for recurrence and malignant progression [1]. Malignant astrocytic tumors such as glioblastoma (GBM) are the most lethal intracranial tumors [2]. The clinical outcome of patients with astrocytic tumors depends on several factors, most notably age at diagnosis, clinical status as measured by the Karnofsky score, and tumor resection extent, as well as the histological classification, tumor grade and key molecular genetic alteration [3]. Despite the treatment of neurosurgical resection, radiotherapy and chemotherapy, most astrocytic tumors still grow continuously and tend to progress to a higher grade, leading to neurological disability and ultimately to death [4]. During recent years, large-scale research efforts – spearheaded by The Cancer Genome Atlas (TCGA) have made rapid advances in understanding glioma genetics. Isocitrate dehydrogenase (IDH) mutations were detected in approximately 80 % of diffuse and anaplastic astrocytomas as well as secondary GBMs [5]. The oncogenic IDH1 mutations mainly represented a change of guanine to adenine at position 395 (G395A), leading to the replacement of arginine by histidine at codon 132 (IDH1-R132H) at the enzymatic active site [5, 6]. Patients with IDH1-R132H positive astrocytic tumors had a better outcome than those with IDH1-R132H negative astrocytic tumors. Recently, several teams demonstrated that IDH mutations and ATRX status, combined with other classical biomarkers, refined the molecular classification of adult gliomas, providing a prognostic tool for clinicians [7–11]. These studies supported the development of a new molecular classification of IDH1-R132H and loss of ATRX, in that the clinical characteristics and prognosis of patients with grade II/III glioma and GBM are not accurately reflected by histological classifications [8, 9, 11–13]. The “Haarlem Consensus Guidelines for Nervous System Tumor Classification” [14] suggest that some entities will require molecular information to provide an “integrated” diagnosis, which is based on several layers comprising (i) the integrated diagnosis as top layer, followed by (ii) histological classification, (iii) WHO grade, and (iv) molecular information [14]. However, the clinical practicability and significance of molecular parameters for the diagnostic and prognostic prediction of astrocytic tumors is still limited. In this study, we detected ATRX, IDH1-R132H and Ki-67 by immunohistochemistry and characterized three prognostic subgroups of astrocytic tumors (referred to as A1, A2 and A3).
RESULTS
Detection of ATRX, IDH1-R132H and Ki-67 in astrocytic tumors by immunohistochemistry
In our dataset, ATRX nuclear protein, IDH1-R132H and Ki-67 was detected by immunohistochemistry. IDH1-R132H dominated in diffuse astroctytoma (29/50, 58%) and anaplastic astrocytoma (5/9, 55.6%) compared with in primary GBMs (9/58, 15.5%) (Figure 1A; Table 1, p<0.0001, Chi-Square test). The frequency of ATRX loss was higher in As (41/50, 82%) and AAs (7/9, 77.8%) than that in pGBMs (7/58, 12.1%) (Figure 1B; Table 1, p<0.0001, Chi-Square test). Primary GBMs (50/58, 86.2%) expressed higher Ki-67 protein than diffuse and anaplastic astrocytoma (1/58, 1.7%) (Figure 1C; Table 1, p<0.0001, Chi-Square test).
Association of IDH1-R132H with ATRX and Ki-67 expression
In accord with previous reports, among the 41 tumor samples with IDH1-R132H positive, 34 lacked ATRX nuclear protein, while 53 samples of 74 astrocytic tumors with IDH1-R132H negative expressed ATRX, indicating a strong association between ATRX loss and IDH1-R132H (Figure 2A; p<0.0001, Chi-Square test). In addition, the Ki-67 high expression restricted in the tumors with IDH1-R132H negative, indicating a group astrocytic tumors with high cell proliferative capacity (Figure 2B; p=0.0129).
Prognostic value of ATRX, IDH1-R132H and Ki-67 for patients with astrocytic tumors
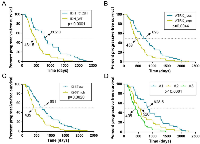
Then we evaluated the value of ATRX, IDH1-R132H and Ki-67 for predicting the progression of astrocytic tumors. We observed that patients with IDH1-R132H positive astrocytic tumors had a longer progressive-free survival (PFS) than those with IDH1-R132H negative astrocytic tumors (Figure 3A; Median PFS of IDH1-R132H positive=802 days, Median PFS of IDH1-R132H negative=461.5 days; p<0.0001). ATRX loss was also a favorable prognostic factor in patients with astrocytic tumors (Figure 3B; Median PFS of ATRX loss=693 days, Median PFS of ATRX expression=459 days; p=0.0044). High Ki-67 expression was associated with shorter PFS in patients with astrocytic tumors (Figure 3C; Median PFS of high Ki-67 expression=405 days, Median PFS of low Ki-67 expression =459 days; p=0.002).
Classification of astrocytic tumors defined by ATRX, IDH1-R132H and Ki-67
Based on the above results and previous research, we incorporated IDH1-R132H, ATRX and Ki-67 status detected by IHC into A1-A2-A3 model. We classified astrocytic tumors into IDH1-R132H positive and IDH1-R132H negative tumors and then defined IDH1-R132H positive and ATRX loss tumors as A1, IDH1-R132H negative tumors with high Ki-67 expression as A3, and grouped IDH1-R132H positive with ATRX expression and IDH1-R132H negative tumors with low Ki-67 expression into A2. We observed that the distribution of patients’ age in the A1-A2-A3 model was significant different (Figure 2C, p=0.003). As showed in Figure 3D, survival analysis of the new classification also demonstrated a remarkable separation of the clinical course in the three molecular subgroups (log-rank test, p<0.0001). The A1 subgroup was correlated with a better clinical outcome (Figure 4, Median PFS = 828.5 days). In contrast, the A3 subgroup was associated with a poorer clinical outcome (Median PFS = 405 days). Correlation of the A2 subgroup with respect to clinical outcome fell between the A1 and A3 subgroups (Median PFS = 526 days). To study the influence of the three molecular markers (IDH1, ATRX and Ki-67) we used in our classification schema, multivariate Cox regression analyses were used for the adjustment of these factors (Table 2). We confirmed that IDH1-R132H status and Ki-67 expression were the independent prognostic factors in this cohort and the new classification scheme was dependent on these three factors to predict survival value. Furthermore, upon incorporation of only our classification and the WHO grading scheme (Table 2), the prognostic value of our classification was still significant, independent of the WHO grades, and served as an addition to the latter.
DISCUSSION
Over the past decade, insights into the molecular pathology of gliomas have significantly improved both our biological understanding of neoplasms as well as our abilities to diagnose tumors and estimate their prognosis and likelihood of response to specific therapies [15]. In the present study, we detected the IDH1-R132H, ATRX and Ki-67 expression using immunohistochemistry, promoting the molecular neuropathology research to the clinical translational practice.
Nowadays, mutations in IDH1 are commonly established as a hallmark molecular feature of grade II/III gliomas and secondary GBM which have predominant localization in the frontal and temporal lobes [15]. IDH1–R132H (G395A) is the most common mutation (~90%), followed at a distance by IDH1– R132S (C394A), IDH1–R132C (C394T), IDH1–R132G (C394G), IDH1–R132L (G395T) and IDH1–R132V (C394G G395T) (0.5–5%) [12]. Thus, IDH1-R132H can be used for the diagnosis between grade II/III gliomas, secondary GBM and primary GBM [16]. Mutations in the IDH genes are thought to cause glioma-CpG island methylator phenotype (G-CIMP) within the proneural GBM subgroup [17]. IDH mutations seem to require cooperating mutations in ATRX, and they are less frequently detected in primary GBMs [5]. Mutations of ATRX inactivated the gene product and caused a lack of ATRX immunolabeling [18]. ATRX loss occurs almost exclusively in IDH mutant astrocytic tumors, and ATRX loss and 1p/19q codeletion are largely mutually exclusive [19]. ATRX loss is characteristic in the refinement of the diagnosis of IDH mutant astrocytomas. Assessment of ATRX loss by immunohistochemical staining captures the majority of mutations, indicating that the use of immunohistochemical testing in routine neuropathology diagnostics gives a reasonable sensitivity [20].
In addition, our result showed that higher Ki-67 expression mostly dominated in the IDH1-R132H negative cluster. Previously, our research delineated that IDH-wt/TERTp-mut gliomas expressed higher Ki-67 protein and showed the evidence of cell proliferation. “Classical” gene expression was mostly restricted to the IDH-wt/TERTp-mut gliomas with the poorest survival. Now, we used negative IDH1-R132H combined with higher Ki-67 expression to define the cluster similar to the IDH-wt/TERTp-mut gliomas. In contrast with IDH mutations and ATRX loss being widely considered as key aberrations in the early stage of astrocytic tumors, higher Ki-67 expression may be the final event in the progression of these tumors. We speculated that IDH1-R132H accompanied by ATRX or Ki-67 may represent a distinct biological process during the development of astrocytic tumors from the original tumor cells.
Based on the above results and previous research, we incorporated IDH1-R132H, ATRX and Ki-67 status detected by IHC into A1-A2-A3 model. The new classification also demonstrated a remarkable separation of the progression interval in the three molecular subgroups and the distribution of patients’ age in the A1-A2-A3 model was also significant different. This model will aid predicting the overall survival and progressive time of astrocytic tumors’ patients.
MATERIALS AND METHODS
Patients enrollment
As a part of the Chinese Glioma Genome Atlas (CGGA) project (http://www.cgga.org.cn/portal.php), we consented patients who underwent surgical resection for malignant gliomas at the Glioma Treatment Center of Beijing Tiantan Hospital from January 2008 through March 2015. The study was approved by the ethics committee in both hospitals and written informed consent was obtained from each patient. All of data and samples were collected under the IRB of Beijing Tiantan Hospital. The criteria of enrollment include: age more than 18 years-old, histologically confirmed astrocytic tumors, relapse detected by MRI and patient’s consent. 117 samples came into the cohort, containing astrocytoma (A, grade II), anaplastic astrocytoma (AA, grade III) and primary glioblastoma (GBM, grade IV). The histological diagnoses were confirmed by two neuropathologists according to the 2007 World Health Organization (WHO) classification guidelines. Specimens were collected after definitive diagnosis and stored as paraffin embedded blocks for subsequent molecular characterization. The collected specimens were verified by our pathologists to harbor >80% viable tumor tissue. For each enrolled patient, patients’ progression-free survival data were recorded when the relapse occurred.
Immunohistochemistry for IDH1-R132H, ATRX and Ki-67
Immunostaining was performed according to the manufacturer’s protocol. In brief, formalin-fixed, paraffin-embedded tissue sections cut to four micrometer were dried at 80 °C for 15 min and dewaxed in xylene, rinsed in graded ethanol, and rehydrated in double-distilled water. The sections were then treated with 3% H2 O2 for 5 min at room temperature (RT) to block endogenous peroxidase activity. For antigen retrieval, slides were pretreated by steaming in sodium citrate buffer (10 mM sodium citrate, pH 6.0) for 15 min at 100°C. After washing with phosphate-buffered saline for 3 min, the sections were immunostained with an anti-human IDH1-R132H antibody (at 1:60 dilution, H09, Dianova, Hamburg, Germany) or an anti-human ATRX antibody (at 1:800 dilution, ab97508, Abcam) or an anti-human Ki-67 protein antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and incubated at 4°C over night. After washed by 3 changes of PBS buffer, the tissues were covered by anti-mouse/rabbit polymer HRP-label for 30min at RT. Staining reaction was performed through covering tissue by prepared DAB chromogen solution, and incubating approximately for 10 min to allow for proper brown color development. Each slide was individually reviewed and scored by two experienced neuropathologists.
Standard of IDH1-R312H staining: (1) a strong cytoplasmic immunoreaction product was scored positive; (2) a weak diffuse staining and staining of macrophages were not scored positive [21, 22].
Standard of ATRX staining according to German Cancer Research Center (DKFZ): nuclear ATRX loss was scored as specific if tumor cell nuclei were unstained while nuclei of non-neoplastic cells such as endothelia, microglia, lymphocytes and reactive astrocytes were strongly positive [12].
Staining was scored using a two-grade scale, with 0=no or <10% occurrence of staining, 1 = >10% of cells positively stained. Score 1 was defined as high Ki-67 expression [5].
Statistical analysis
Receiver operating characteristic (ROC) curves were constructed to determine the discriminatory capacity of IDH1-R132H and (or) ATRX loss for diagnosis. Kaplan-Meier analysis was performed to estimate the survival time of different subgroups and a log-rank test was used to test prognostic differences. Comparisons of binary and categorical patient characteristics between subgroups were performed by the use of the Fisher’s exact test All statistical computations were performed with the statistical software environment R version 3.2.0 ( http://www.r-project.org/), GraphPad Prism Version 6.01 or Microsoft Excel 2013. P value < 0.05 was considered statistically significant.
ACKNOWLEDGMENT
We thank Yuling Yang for tissue sample collection and clinical data retrieval. This work was supported by grants from The Research Special Fund For Public Welfare Industry of Heath (No. 201402008), The National Key Research and Development Plan (No. 2016YFC0902500). National Key Technology Research and Development Program of the Ministry of Science and Technology of China (No. 2014BAI04B02), National High Technology Research and Development Program (No.2012AA02A508), Beijing Science and Technology Plan (No.Z131100006113018), International Science and Technology Cooperation Program (No. 2012DFA30470), National Natural Science Foundation of China (No.81372700,No. 81201993), Special Fund Project of Translational Medicine in the Chinese-Russian Medical Research Center (No. CR201417), Research Project of Chinese Society of Neuro-oncology, CACA (CSNO-2014-MSD08).
CONFLICTS OF INTEREST
None of the authors has a conflicts of interest regarding this study.


- 1. Loss of ATRX, associated with DNA methylation pattern of chromosome end, impacted biological behaviors of astrocytic tumors. Oncotarget. 2015; 6:18105-18115. https://doi.org/10.18632/oncotarget.3906. [PubMed].
- 2. IDH mutation and MGMT promoter methylation in glioblastoma: results of a prospective registry. Oncotarget. 2015; 6:40896-40906. https://doi.org/10.18632/oncotarget.5683. [PubMed].
- 3. Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology : official journal of the European Society for Medical Oncology /ESMO. 2014; 25 Suppl 3:iii93-101. https://doi.org/10.1093/annonc/mdu050. [PubMed].
- 4. Recurrence and malignant degeneration after resection of adult hemispheric low-grade gliomas. Journal of neurosurgery. 2010; 112:10-17. https://doi.org/10.3171/2008.10.JNS08608. [PubMed].
- 5. ATRX mRNA expression combined with IDH1/2 mutational status and Ki-67 expression refines the molecular classification of astrocytic tumors: evidence from the whole transcriptome sequencing of 169 samples samples. Oncotarget. 2014; 5:2551-2561. https://doi.org/10.18632/oncotarget.1838. [PubMed].
- 6. IDH Mutation in Glioma: New Insights and Promises for the Future. JAMA neurology. 2014. https://doi.org/10.1001/jamaneurol.2014.1205. [PubMed].
- 7. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2015. https://doi.org/10.1038/ng.3457. [PubMed].
- 8. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine. 2015. https://doi.org/10.1056/NEJMoa1402121. [PubMed].
- 9. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. The New England journal of medicine. 2015. https://doi.org/10.1056/NEJMoa1407279. [PubMed].
- 10. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015; 28:318-328. https://doi.org/10.1016/j.ccell.2015.07.013. [PubMed].
- 11. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015. https://doi.org/10.1038/ng.3273. [PubMed].
- 12. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015; 129:133-146. https://doi.org/10.1007/s00401-014-1370-3. [PubMed].
- 13. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget. 2015. https://doi.org/10.18632/oncotarget.4497. [PubMed].
- 14. Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, Figarella-Branger D, Fuller GN, Giangaspero F, Giannini C, Hawkins C, Kleihues P, et al. International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain pathology (Zurich, Switzerland). 2014; 24:429-435.
- 15. Evolving Molecular Genetics of Glioblastoma. Chinese medical journal. 2016; 129:464. https://doi.org/10.4103/0366-6999.176065. [PubMed].
- 16. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Scientific reports. 2015; 5:16238. https://doi.org/10.1038/srep16238. [PubMed].
- 17. Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, MelnickA, Wellen KE, O'Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012; 483:474-478. https://doi.org/10.7554/eLife.10860. [PubMed].
- 18. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011; 333:425. https://doi.org/10.1126/science.1207313. [PubMed].
- 19. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013; 126:443-451. https://doi.org/10.1007/s00401-013-1156-z. [PubMed].
- 20. Review: Diagnostic, prog-nostic and predictive relevance of molecular markers in gliomas. Neuropathology and applied neurobiology. 2015. https://doi.org/10.1111/nan.12246. [PubMed].
- 21. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain pathology (Zurich, Switzerland). 2010; 20:245-254. https://doi.org/10.1111/j.1750-3639.2009.00352.x. [PubMed].
- 22. Cai, J., et al. (2016). “Detection of ATRX and IDH1-R132H immunohistochemistry in the progression of 211 paired gliomas.” Oncotarget 7:16384-16395. https://doi.org/10.18632/oncotarget.7650. [PubMed].
Last Modified: 2016-09-12 11:56:06 EDT
