Oncoscience
Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following Cetuximab treatment: biomolecular, genetic and translational implications
Marco Ragusa1,*, Luisa Statello1,*, Marco Maugeri1,*, Cristina Barbagallo1, Roberta Passanisi1, Mohamed S. Alhamdani2, Giovanni Li Destri3, Alessandro Cappellani4, Davide Barbagallo1, Marina Scalia1, Hadi Valadi5, Jörg D. Hoheisel2,**, Cinzia Di Pietro1,**, Michele Purrello1,**
1 Molecular, Systems and Genome BioMedicine Unit, Department Gian Filippo Ingrassia, University of Catania, Catania, Italy, EU
2 Division of Functional Genome Analysis, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany, EU
3 Dipartimento di Scienze Chirurgiche, Trapianti d’Organo e Tecnologie Avanzate, Università di Catania, Catania, Italy, EU
4 Dipartimento di Chirurgia, Università di Catania, Catania, Italy, EU
5 University of Gothenburg, Department of Rheumatology and Inflammation Research, Gothenburg, Sweden, EU
* Equal Contribution. ** Senior Corresponding Authors.
Correspondence to: Michele Purrello, email: [email protected]
Keywords: Cetuximab, Colon Cancer, Exosomes, miRNAs, Proteins
Received: February 24, 2014
Accepted: March 14, 2014
Published: March 16, 2014
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT:
Exchange of molecules via exosomes is a means of eukaryotic intercellular communication, especially within tumour microenvironments. However, no data are available on alterations of exosomal molecular cargo by environmental cues (eg, pharmacological treatments). To approach this issue, we compared the abundance of 754 miRNAs and 741 cancer-related proteins in exosomes secreted by Caco-2 (Cetuximab-responsive) and HCT-116 (Cetuximab-resistant) CRC cells, before and after Cetuximab treatment, with that in their source cells. Cetuximab significantly altered the cargo of Caco-2 exosomes: it increased abundance of miRNAs and proteins activating proliferation and inflammation and reduced miRNAs and proteins related to immune suppression. These alterations did not precisely mirror those in source cells, suggesting a Cetuximab-linked effect. Analogous alterations were detected in HCT-116. Transfection of exosomes from Cetuximab-treated Caco-2 into HCT-116 significantly increased HCT-116 viability; conversely, no viability alteration was detected in Caco-2 transfected with exosomes from Cetuximab-treated HCT-116. Analysis of networks, comprising targets of differentially expressed (DE) exosomal miRNAs and DE exosomal proteins, demonstrates a significant involvement of processes related to proliferation, inflammation, immune response, apoptosis. Our data extend existing knowledge on molecular mechanisms of eukaryotic intercellular communication, especially in oncological processes. Their translation to clinical settings may add new weapons to existing therapeutic repertoires against cancer.
INTRODUCTION
Exosomes are nanosized vesicles(50-120 nm in diameter), which derive from endosomal compartment invaginations called Multivesicular Bodies (MBVs). MBVs may be released into the extracellular milieu by fusion with the plasma membrane as an alternative to fusion with the lysosomal compartment [1, 2]. Exosomes are synthesized by possibly all cell phenotypes, including tumour cells and platelets [3 - 6]. They also have been found in several body fluids (eg, blood, saliva, urine, bronchoalveolar fluid, synovial fluid, amniotic fluid, breast milk, malignant ascite) [7 - 12]. Exosome internalization by recipient cells follows binding of exosomal membrane adhesion molecules to cellular receptors (eg, LFA1, TIM1, TIM4) [13, 14]. Exosomes from different cell phenotypes and biological fluids have been shown to contain functional subsets of proteins, mRNAs and miRNAs, which can be transferred to recipient cells: this would suggest a novel and still partially unexplored mechanism of communication and genetic exchange among eukaryotic cells [15 - 19]. Sorting of RNAs and proteins into exosomes is likely a selective process, whose general rules and signals started to be clarified only recently [15, 20, 21]. Notably, the RNA cargo of exosomes may partially diverge from that of source cells: in fact, the same cells if cultured under different conditions (eg, serum starvation, oxidative stress) may produce exosomes with different molecular cargos [15, 22]. Accordingly, it would seem logical to hypothesize that nucleic acids and proteins may be selectively sorted into exosomes depending on the specific molecular signals they carry. Exosomes have been shown to be involved in different biological functions, as intercellular communication, modulation of immune functions, transport and propagation of prions and retroviruses [23 - 25]. Tumour-derived exosomes have been found to possess immunosuppressive and protumorigenic properties, since they apparently facilitate tumour growth and metastasis [6, 26]. It has been recently noted that tumour cells possess increased exosomes-shedding properties compared to normal cells [27]. Significantly increased levels of serum exosomes were detected in patients with ovarian cancer, compared to healthy controls [28]. Also, high levels of exosomes in plasma of colorectal cancer (CRC) patients were secreted by poorly differentiated tumours and were associated with decreased overall survival [29]. Despite these scientific advances, the biologic and pathologic significance of exosomes is still not fully understood. Based on these premises, we sought to analyze the alterations of exosomal miRNAs and proteins cargo profiles from CRC cells following Cetuximab treatment. Cetuximab (Erbitux) is a monoclonal antibody that acts as competitor of natural ligands of the Epidermal Growth Factor Receptor (EGFR): Epidermal Growth Factor, Amphiregulin, Epiregulin, Transforming Growth Factor α. Accordingly, it is among the most commonly used drugs for treating advanced metastatic CRC [30]. Cetuximab binding to EGFR impairs its activation and leads to a block of downstream KRAS proliferative signalling, thus affecting tumour cells proliferation [30]. However, KRAS mutations in CRC patients may lead to permanently activated EGFR pathway regardless of the EGFR status, so preventing efficacy of Cetuximab-based therapy [31]. In previous work, we have shown that in vitro sensitivity of CRC cells to Cetuximab depends on specific miRNA transcriptome profiles [32]. Interestingly, a correlation between exosomes and effectiveness of monoclonal antibody-based therapy has already been found in breast cancer: exosomes secreted by HER2-overexpressing breast carcinoma cells express full-length HER2 molecules on their surface, which bind and sequester Trastuzumab (a therapeutic monoclonal antibody) and lower its therapeutic efficacy [33]. The data reported in this paper demonstrate that Cetuximab significantly alters the miRNAs and proteins cargo of exosomes released by CRC cells. Intriguingly, we also show that transfection of steady-state or Cetuximab-treated HCT-116 (Cetuximab unresponsive) with exosomes from Cetuximab-treated Caco-2 (Cetuximab sensitive) significantly increases HCT-116 viability and alters their Cetuximab responsiveness.
RESULTS
Characterization of Exosomes from Caco2 and HCT-116 cells
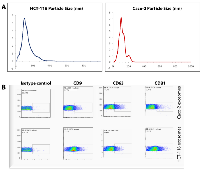
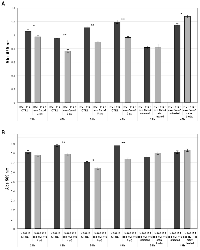
Following exosome isolation, the size of pelleted particles was determined with dynamic light scattering using a Zetasizer Nano. The results showed that the pellet consisted of particles with an average size of 100 nm in diameter: this is consistent with the characteristic size range of exosomes (Figure 1A). By flow cytometry, we confirmed that the isolated nanostructures stained positive for canonical exosome markers CD9, CD63 and CD81 (Figure 1B).
Profiling of exosomal and cellular miRNAs before and after Cetuximab treatment
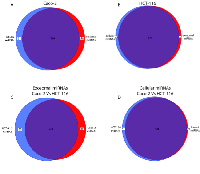
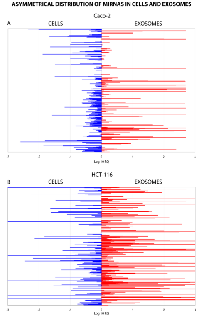
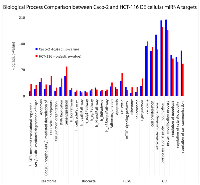
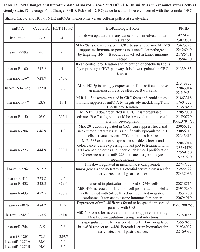
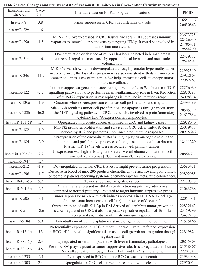
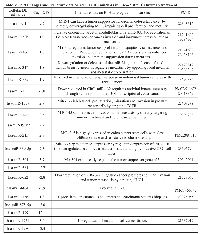
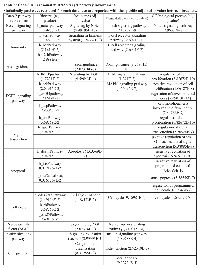
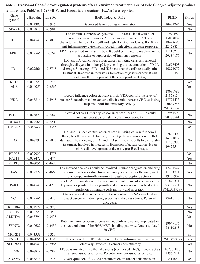
Using TaqMan Low Density Array (TLDA) technology, we determined the expression profiles of 754 miRNAs in exosomes from Caco-2 and HCT-116 cells; through the analysis of the same samples, we also characterized the miRNA content in the respective source cells. In all cases, analysis was performed before and after seven days of Cetuximab treatment. We compared the sets of steady-state miRNAs in Caco2 cells (447 molecules detected), Caco2 exosomes (430), HCT-116 cells (469), and HCT-116 exosomes (466) (Figure 2). Both cell lines reciprocally shared about 93% of cellular miRNA species and about 90% exosomal miRNA species (Figure 2, C and D). Opposed to the overlap between exosomal and cellular miRNAs in the qualitative analysis, we detected a strong asymmetric distribution of miRNAs between secreted exosomes and their source cells when we subjected our data to quantitative analysis (Figure 3, A and B). Intriguingly, some miRNAs were found to be specifically located in exosomes (eg, miR-127, miR-136*, miR-144, miR-432, miR-433, miR-487b and miR-495 in Caco-2 exosomes; miR-136*, miR-223*, miR-380-5p, miR-432 and miR-672 in HCT-116 exosomes) (Figure 2, A and B). Steady-state exosomes from both cell lines were highly enriched in miRNAs involved in blocking proliferation and immune escape (eg, miR-142-5p, miR-150, miR-223, miR-433) (Table 1). Profiling of exosomal miRNAs after Cetuximab treatment showed changes for 25 and 20 miRNAs in Caco2 exosomes and source cells, respectively (Table 2 and 3); in comparison, we detected 9 and 12 miRNAs whose levels had been altered by Cetuximab in HCT-116 exosomes and source cells, respectively (Table 4). Notably, the steady-state asymmetric distribution of miRNA content in exosomes and their source cells was highly accentuated by Cetuximab treatment. Indeed, the sets of DE miRNAs in exosomes and source cells lines did not overlap qualitatively for both cell lines, with the exception of miR-31* that was moderately upregulated both in treated Caco-2 cells and their exosomes. Specifically, many DE miRNAs in the exosomes secreted by Caco2 also are involved in proinflammatory mechanisms in addition to performing cancer-related functions (eg, let-7a, miR-122, miR-133b, miR-511) (Table 2). In Cetuximab-treated Caco2 cells, we also detected upregulation of the CRC tumour-suppressors miR-1, miR-133a, miR-145 and miR-215 (Table 3). Considering both the number of DE miRNAs and the magnitude of changes, exosomal miRNA alterations in KRAS-mutated cells (HCT-116) were less pronounced (Table 4). Noteworthy was the downregulation of miR-624 and miR-1289, which are orphans of functions in CRC biopathology. Highly downregulated was also miR-802, a miRNA reported to perform a tumour suppressor role in intestinal epithelial cells (Table 4). The set of DE cytoplasmatic miRNAs in HCT-116 cells was characterized by moderate upregulation of several putative oncomirs (eg, miR-193a-3p, miR-424*, miR-501-3p, miR-938, miR-1285) (Table 4).
Gene Ontology analysis of exosomal and cellular miRNAs in CRC cells before and after Cetuximab treatment
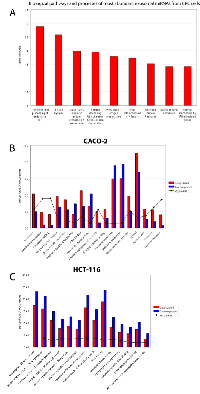
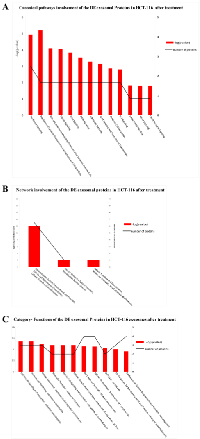
To infer the biological processes regulated by exosomal and cellular DE miRNAs at steady-state and after Cetuximab treatment, we retrieved validated and predicted miRNA targets; then, we statistically analysed their Gene Ontologies and pathways involvement by David and FatiGo tools. According to Reactome Database, targets of exosomal miRNAs from both cell lines at steady-state were statistically enriched in genes and proteins involved in modulation of the immune system (trafficking and processing of endosomal Toll-like receptors, antigen processing and presentation, unfolded protein response) (Figure 4A). This agrees with literature reports that the most abundant miRNAs in steady-state CRC exosomes are mainly involved in immune escape (Table 1). For treated Caco-2 cells, exosomal DE miRNAs were functionally enriched in biological processes as immunity, EGFR pathway, apoptosis, cell cycle (Table 5). Upregulated miRNAs were statistically more present in molecular processes associated with negative control of translation than those downregulated (Figure 4B). For treated HCT-116, exosomal DE miRNAs were enriched in biological processes related to apoptosis, cell cycle and immunity (Table 6). On the other hand, downregulated miRNAs showed a significant overabundance of GO terms related to transcription, positive regulation of transcription and positive regulation of metabolic processes when compared to overrepresented miRNAs (Figure 4C). Direct comparison of GO categories and pathways of the targets of DE miRNAs after Cetuximab treatment of Caco-2 and HCT-116 cells, showed several statistically significant differences: targets of DE miRNAs from Caco-2 cells were more represented in intracellular signalling and cell communication than those from HCT-116 cells; in HCT-116 cells, we observed a statistically significant expression of cell cycle and gene expression processes when compared to Caco-2 cells (Figure 5).
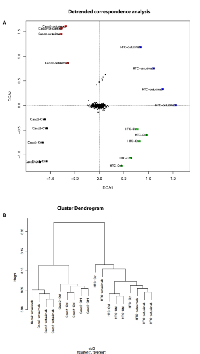
Profiling of exosomal proteins before and after Cetuximab treatment
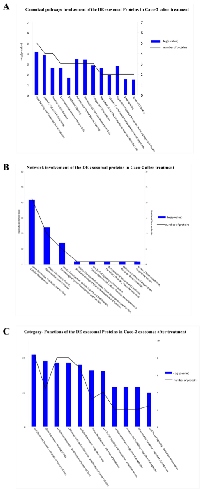
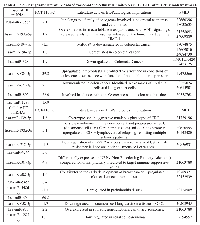
Using antibody microarrays, we profiled the expression of 741 cancer-related exosomal proteins in Caco-2 and HCT-116, at steady-state and after Cetuximab treatment. For this analysis, 18 biological replicates were selected on the basis of their concentration, 5 controls and 4 treated samples each for both cell lines. Similarities and differences among the various sample groups were globally assessed using Hierarchical Clustering, Non-Metric Multidimensional Scaling, and Detrended Correspondence Analysis (Figure 6A). For both cell lines, treated samples and controls showed a unique and reproducible protein expression pattern. Normalized data are presented in a cluster dendrogram tree (Figure 6B). Statistically significant DE exosomal proteins for both cell lines are shown in Tables 7, 8 and 9. We found 54 exosomal DE proteins in Caco-2 compared to controls (27 overexpressed, 27 downregulated), and 9 exosomal DE proteins in HCT-116 (5 overexpressed and 4 downregulated). These data indicate different qualitative alterations of exosomal proteomes of the two cell lines, which exhibit different sensitivity to Cetuximab. Only VCAM1 (an important protooncoprotein involved in angiogenesis) was overexpressed in exosomes from both cell lines following treatment. The strongest overexpressed proteins in treated Caco-2 exosomes (eg, CD59, CRP, CTSD, HMMR, TRIM22) are involved as potential oncogenes in the pathogenesis of different cancers, including CRC (Table 8). In addition, some important protooncogenes involved in cell cycle control (eg, BRAF, CDC2, ERBB2) were moderately overexpressed. Among the most downregulated proteins, we found several confirmed or candidate tumour suppressor genes (eg, ID1, FAS, FPR1, IL10, PCGF2, VDR). Notably, some exosomal DE proteins are components either of intracellular macromolecular complexes or of organelles (eg, ribosomes: RPL7, RPL10A, RPS15; mitochondria: MRPL3, ATP5H). Most overexpressed proteins are involved in immune response, as T-cells activation and functions (CD59, IL12A) or the response to bacterial and viral infections (CTSD, TRIM22). Opposed to this, some downregulated proteins are involved in immune suppression through induction of apoptosis and antiinflammatory activity (eg, EP300, FAS, IL10). Important to stress, only about 50% of Caco-2 DE proteins were already annotated in Exocarta (http://www.exocarta.org/), a comprehensive database of human exosomal proteins; for all other proteins, this is the first report on their presence in exosomes, in particular for CRC cells. The low number of DE exosomal proteins in treated HCT-116 suggests that Cetuximab altered their exosomal proteome to a lower extent than in Caco-2 (Table 9); this could be a hint to the molecular basis of HCT-116 resistance to treatment. Among downregulated proteins, PTEN is a well known tumour suppressor in CRC, while CDC20, TNF and VCAM1 are important protooncogenes. Four DE proteins are involved in immunity and intercellular signalling (IL1B, MS4A2, PTEN, TNF) (Table 9).
Network and canonical pathway analysis of exosomal proteins in CRC cells before and after Cetuximab treatment
DE exosomal proteins were analyzed to determine their GO classification and pathway involvement through Ingenuity software. Network and canonical pathway analysis for Caco-2 exosomal DE proteins yielded particularly relevant results, as they showed their important involvement in cell growth and proliferation, intercellular signalling, and cell death (Figure 8, A, B, C); also well represented are the pathways related to immune response (ie, IL-12 signalling and production in macrophages, T- and B-cell signalling in immune diseases, T helper cell differentiation and activation of leukocytes). The same analysis for HCT-116 exosomes demonstrated that most DE proteins are involved in biological processes as cell to cell signalling, cell growth and proliferation, cell death, and cell movement (Figure 7 A and B). A small number of DE proteins is involved in immune cell trafficking and inflammatory response (Figure 7C).
Modifications of cell viability following exosomes transfection
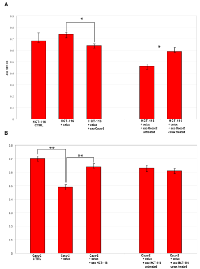
To analyze the biomolecular effects of exosomes secreted by CRC cells before and after Cetuximab treatment, we transfected HCT-116 cells with exosomes from steady-state or Cetuximab-treated Caco-2 cells and checked HCT-116 viability at 24 h and 48 h after transfection (AT). In turn, Caco-2 cells were transfected with steady-state or Cetuximab-treated HCT-116 exosomes and analyzed the same way. HCT-116 treated with 2 µg of steady-state Caco-2 exosomes showed a slight decrease of viability at 24 h AT, which became more evident at 48 h AT. By using 5 µg of exosomes, the differences of viability were even more pronounced (Figure 9A). Also Caco-2 cells that had been incubated with HCT-116 exosomes exhibited significantly lower proliferation rates at 48 h AT (Figure 9B). When HCT-116 cells were transfected with exosomes from treated Caco-2, we observed no significant variation at 24 h AT; proliferation rates increased significantly at 48 h AT in comparison to HCT-116 cells transfected with exosomes from untreated Caco-2 (Figure 9A). Caco-2 cells transfected with exosomes from treated HCT-116 showed no significant difference in cell viability compared to controls (Figure 9B). Treatment with Cetuximab of HCT-116 cells, which had been transfected with 5 µg of steady-state exosomes, decreased their viability (Figure 10A). In contrast, Caco-2 cells simultaneously incubated with Cetuximab and exosomes from steady-state HCT-116 increased their viability compared to Caco-2 cells, which had been treated only with Cetuximab (Figure 10B). Notably, cell proliferation increased when we administered Cetuximab to HCT-116 cells incubated with exosomes from Cetuximab-treated Caco-2 cells (Figure 10A). We found no appreciable difference of viability when Caco-2 cells were treated with Cetuximab and transfected with exosomes purified from treated HCT-116 cells (Figure 10B). Overall, these data suggest that exosomes from untreated cells decreased cell viability, when incubated with recipient cells different from their source cells. In constrast, exosomes from Cetuximab-treated cells induced a proliferation increase only when purified from Caco-2 cells, but not from HCT-116. It is worth to stress that simultaneous incubation with Cetuximab and exosomes altered viability of both cell lines (Figure 10A and B).
DISCUSSION
Molecular cargo of exosomes from Caco2 and HCT-116 at steady-state
When they were first discovered, exosomes were considered garbage bags used by cells to get rid of unneeded or dangerous molecules. However, following the characterization in the mid-1990s of extracellular vesicles from antigen-presenting lymphocytes, exosomes were associated with immune system functions [34 – 36]. In recent years, many reports have convincingly demonstrated an important function of exosomes: they work as shuttles transporting signalling molecules (eg, mRNAs, miRNAs, proteins) involved in intercellular communication [15]. Intercellular exchange of nucleic acids and proteins via exosomes has been shown to be an effective mechanism of intercellular communication, especially within the tumor microenvironment [37]. Indeed, transport of RNAs and proteins from tumor cells to neighbouring cells and vice versa could have significant effects on their molecular phenotype. For instance, in tumorigenesis it could modulate proliferation, invasion and cell immunoreactivity. An important protumorigenic role could be performed by tumor-derived exosomes through their involvement in drug resistance: (1) exosome secretion could be utilized by cancer cells to expel anticancer drugs; (2) surface molecules from cancer-derived exosomes could compete for binding with antibody-based drugs, so lowering their therapeutic efficacy [33, 38]. Quite surprisingly, no data have been published to date on an important biological and translational issue: could the molecular composition of exosomal cargo be modulated by drug treatment? In this work, we have demonstrated significant alterations of both exosomal miRNAs and oncoproteins cargo in CRC cells following treatment with anti-EGFR antibodies. These molecular alterations did not precisely mirror the changes that occurred at the same time in source cells. Specifically, we analyzed exosomal miRNAs and proteins from Cetuximab-treated Caco-2 (sensitive) and HCT-116 (resistant) cells. Indeed, more than 90% of miRNAs expressed at steady-state conditions were shared by exosomes and their source cells for both cell lines. However, a sizeable fraction of miRNAs was exosome-specific. Most important, the asymmetric quantitative distribution of miRNAs between exosomes and cells, already present in both cell lines at steady-state, was significantly increased by treatment with Cetuximab. Interestingly, exosomes from both lines shared miRNAs whose amount was highly altered in comparison to source cells. Several upregulated exosomal miRNAs had been previously reported to be differentially expressed in CRC (eg, miR-144*, miR-150, miR-204, miR-411, miR-487b); some of them have potential immunosuppressive activity on T- and B-cells (eg, miR-142-5p, miR-144*, miR-150, miR-433). Extracellular miR-144* was found to be overexpressed in feces of CRC patients, suggesting that it could be a candidate diagnostic marker for non-invasive tumour detection [39]; this miRNA also could be involved in regulating antituberculosis immunity through modification of cytokine synthesis and reduced proliferation of T-cells [40]. Expression levels of miR-150 decreased during progression of CRC tumors [41]; its enforced expression in colon HT29 cells lowered c-Myb and Bcl-2 levels, thus enhancing cell apoptosis [42]. MiR-150 is upregulated in mature resting B- and T-cells, while it is strongly downregulated in their cell precursors and upon immune activation [43]; its overexpression in hematopoietic stem and progenitor cells impairs B-cell and T-cell development [44]. MiR-433 inhibits HeLa cell proliferation following treatment with 5-FU; it is downregulated in gastric carcinoma [45, 46]; it also reduces NK cell cytotoxicity and acts synergistically with other miRNAs to allow immune control escape [47]. Gastric cancer patients with high frequency of recurrence and poor survival outlook showed low levels of miR-142-5p [48]; in contrast, its expression is elevated in peripheral blood leukocytes from IL-10(-/-) mice during minimal colon inflammation [49]; inhibiting miR-142-5p in healthy donor CD4+ T-cells caused T-cell overactivation and B-cell hyperstimulation, whereas its overexpression in Systemic Lupus Erythematosus CD4+ T cells had the opposite effect [50]. Taken together, these data suggest that most upregulated miRNAs in exosomes from Caco-2 and HCT-116 cells show a dual biological function: (i) negative regulation of proliferation or apoptosis induction in cancer cells; (ii) immunosuppression in B- and T- cells. This agrees with our biologic data showing a decreased proliferation of Caco-2 cells after transfection with HCT-116 derived exosomes and vice versa. This anticancer effect of tumor-derived exosomes on tumor cells was already reported for other tumors, but not for CRC: exosomes secreted by human pancreatic tumor cells increased Bax and decreased Bcl-2 expression, inducing the mitochondrial apoptotic pathway in pancreatic cancer cells [51]. Moreover, these exosomes also decreased expression of the intranuclear target of Notch-1 signalling pathway and thereby negatively modulated its prosurvival functions [51]. This somehow unexpected proapoptotic role of cancer-derived exosomes could be explained by hypothesizing a negative autoregolatory loop of their own growth: tumors could exploit this strategy to favor the recruitment of endothelial cells and the establishment of neovasculature. Assuming that the functions of CRC exosomes, which we observed in vitro, mimic those occurring in vivo, it may be hypothesized that apoptotic and antiproliferative activities of exosomes also are addressed to immune cells to escape the immune system, in addition to cancer cells to regulate the tumor growth rate [51]. Notably, our GO analysis on targets of exosomal miRNA from both cell lines at steady-state showed a statistical enrichment in proteins involved in the modulation of the immune system. Diverse immunosuppressive effects of tumor-derived exosomes have been reported. Tumor-derived exosomes (including those from CRC cells) were shown to directly suppress the activity of effector T-cells by activating the expression of death ligands (eg, FasL, TRAIL), which can trigger the apoptotic death of activated T-cells [52, 53]. Interestingly, when CRC cells were treated with Cetuximab and incubated with exosomes from untreated cells, we observed an alteration of their viability. Specifically, cell vitality decreased slightly in Cetuximab-treated HCT-116 cells incubated with Caco-2 exosomes; in contrast, we observed an increase of viability in Cetuximab-treated Caco-2 cells incubated with HCT-116 exosomes. This latter phenomenon is biologically consistent with results reported by Ciravolo et al. and Battke et al. on Trastuzumab-treated breast carcinoma cells [33, 54]: exosomes secreted by HER2-overexpressing breast carcinoma cell lines express HER2 molecule, enabling them to bind to HER2 antibody Trastuzumab both in vitro and in vivo; these exosomes efficiently bound and sequestered tumor-reactive antibodies and dramatically reduced their binding to tumor cells: in this way, they inhibited the overall effect of Trastuzumab on cancer cells proliferation. Given that exosomes express different forms of EGFR, the molecular target of Cetuximab [55 - 57], extracellular binding between Cetuximab and exosomal EGFR could reduce antibody binding to CRC cancer cells, partially neutralizing the effects of antibody-based therapeutics.
Exosomes molecular cargo in Caco2 and HCT-116 cells after Cetuximab treatment
Exosomes represent a vesicle-based mechanism of cancer cells to signal changes to the microenvironment of a developing tumor. Accordingly, it is not surprising that pharmacologically-treated cancer cells alter the content of molecular signals carried inside the exosomes. Indeed, our data demonstrate that the asymmetric expression of miRNAs in exosomes and cytoplasm, already existing at steady-state, was significantly accentuated after Cetuximab treatment: exosomal miRNAs differentially expressed after treatment did not overlap with those detected in source cells. These data suggest that miRNAs alterations inside the exosomes were not due to an osmotic effect of expression variation of the cytoplasmic miRnome, but they were specifically induced in the nanovesicles. More specifically, we found 25 DE miRNAs in exosomes from treated Caco-2 cells involved as oncomirs or tumour suppressors in many cancer-related functions. Intriguingly, most dysregulated miRNAs (let-7a, miR-122, miR-133b, miR-511) had been already reported to participate in inflammatory processes. Let-7a negatively controls IL-13, a cytokine essential for allergic lung disease, an experimental model of asthma [58]. In liver, miR-122 deficiency causes inflammation and fibrosis that lead to hepatocarcinogenesis [59]. A tight coregulation with the miR-206/133b cluster induces proinflammatory cytokine IL-17A in lymphocytes; moreover, miR-133b was found in inflammatory microvesicles in association with metabolic and cardiovascular diseases [60, 61]. MiR-511 is a putative positive regulator of Toll-like receptor 4, an important initiator of native immune responses and inflammation [62]. Interestingly, our data on exosomal protein profiles from treated Caco-2 showed an enrichment of upregulated proteins involved in tumor etiopathogenesis, including CRC (CD59, CRP, CTSD, HMMR, LY6K, TRIM22), and in cell cycle control (BRAF, CDC2, ERBB2). However, some of these proteins also are involved in immune response and inflammation, as T-cells function and activation (CD59, IL12A) [63, 64]; others are involved in the response to bacterial and viral infections (CTSD, TRIM22) [65, 66]. On the other hand, we found FAS, FPR1, ID1, IL10, PCGF2, VDR among downregulated proteins: all are involved in tumor immune-escape through induction of apoptosis and anti-inflammatory activities [67]. Not surprisingly, analysis of networks and canonical pathways of exosomal DE proteins in treated Caco-2 cells showed again a general association of these proteins with the immune response (eg, IL-12 signalling and production in macrophages, T- and B-cell signalling in immune diseases, T helper cell differentiation, activation of leukocytes). Taken together, these data suggest that the molecular exosomal cargo (miRNAs and proteins), secreted by Caco-2 after treatment with Cetuximab, mainly carry proinflammatory signals. This observation on exosomes from Cetuximab-sensitive cells is potentially important, considering that response to this drug is often associated with immune activation and inflammation in CRC patients. In previous work, it was reported that most of CRC cellular DE miRNAs at 24 and 48 h after treatment controlled targets related to immune activation [32, 68]. Moreover, several studies have shown that in tumor microenvironment inflammation contributes to proliferation and survival of malignant cells, angiogenesis, metastasis, subversion of adaptive immunity, reduced response to hormones and chemotherapeutic agents [69 - 71]. Accordingly, exosomes could contribute with their miRNAs and proteins to induce in vivo proinflammatory phenomena in CRC treated patients; these would result in a protumoral behaviour. This latter hypothesis is supported by our results on significantly increased cell viability of HCT-116 (at steady-state and after Cetuximab treatment) after incubation with exosomes from treated Caco-2 cells. As already reported in the Results section, miRNAs and proteins alterations in exosomes from Cetuximab-treated HCT-116 were less striking than those detected in Caco-2, both for the number of DE molecules as for the magnitude of the changes. This difference could be due to the Cetuximab resistance of these cells. Indeed, we observed no relevant variation of cell viability when Caco-2 cells were incubated with exosomes from treated HCT-116 cells.
CONCLUSIONS: BIOMOLECULAR, GENETIC AND TRANSLATIONAL IMPLICATIONS
During tumor progression, cancer cells modulate their molecular phenotype through genetic and epigenetic mechanisms [72, 73]. These alterations also involve qualitative and quantitative changes of the exosomal cargo, which in turn affect the microenvironment (this paper and cited bibliography). The predominant biomolecular functions of exosomes depend on their specific molecular phenotype as on that of their source cells. In addition, environmental factors seem to have an important role in determining the behaviour and immunologic impact of tumor-derived exosomes. When pharmacologically treated, CRC cells specifically alter miRNAs and proteins exosomal profiles. The overall biological effects of these alterations are related to in vitro drug resistance and tumor survival, but they also are likely to reflect in vivo phenomena occurring in patients. Careful in vivo molecular analysis of serum exosomes from large-scale epidemiological studies on cohorts of CRC patients before and after Cetuximab treatment is warranted before clinical applications. Intriguingly, knowledge of how exosomes could influence the tumour environment in vivo during therapy could pave the way to develop procedures of exosome block or removal as an additional therapeutic strategy in CRC patients [74]. Switching to a wider scientific horizon, our results and those from the literature also confirm that important genetic and biologic features could be horizontally transferred among eukaryotic cells through exosomes and their molecular cargo: these potentially paradigm-changing perspectives should be carefully investigated [75, 76].
MATERIALS AND METHODS
Cell lines
Caco-2 and HCT-116 cells are from the Interlab Cell Line Collection (ICLC), an international Repository Authority within the IRCCS Azienda Ospedaliera Universitaria San Martino-IST Istituto Nazionale per la Ricerca sul Cancro (Genova, Italy, EU). Caco-2 cells were cultured in Eagle minimal essential medium (EMEM) (Cambrex Bio Science), supplemented with 20% fetal bovine serum (FBS), 2 mM L-glutamine, 1% NEAA (Gibco); HCT-116 cells were cultured in McCoy 5A medium (Gibco), supplemented with 10% FBS and 2 mM L-glutamine (Gibco). Characterization and validation of cell lines were performed by the cell repository: cell lines were verified to be mycoplasma-free by Hoechst staining and PCR (TIB Molbiol), and by MycoTect (Gibco BRL); species verification was performed by isoenzyme analysis (AuthentiKit TM System, Innovative Chemistry): multiplex short-tandem-repeat profiling confirmed identity and uniqueness of cell lines. After receiving cells from ICLC, an aliquot was cultured up to the 10th passage to perform the experiments; remaining cells were immediately frozen at -196°C. FBS was depleted of exosomes by centrifugation for 70’ at 120,000 g followed by filtration through 0.2 µm filters.
Cetuximab treatment
A total of 5 x 106 or 6 x 106 cells were seeded in 175 cm2 culture flasks in serum starvation conditions (1% FBS) for CaCo-2 and HCT-116, respectively. Cells were cultured for seven days and treated with 40 µg/mL Cetuximab (Erbitux®, Merck KGaA, Darmstadt, Germany) each second day. On the seventh day of treatment (PT), exosomes were isolated from culture media (see below). RNAs and proteins were extracted from the same aliquots of exosome preparations as from the same aliquots of source cells, for both treated and untreated samples (the latter were treated with an equivalent volume of PBS, the solvent of Cetuximab). All experiments were performed in biological triplicates.
Exosome isolation, characterization and transfection
Exosomes were extracted from cell culture supernatants by centrifugation at 300 g to pellet debris, and then at 16,500 g for 30’, followed by filtration through a 0.2 µm filter. The final supernatant was ultracentrifuged at 120,000 g on a Beckman L8-70M ultracentrifuge in a SW28 rotor for 70’. Exosome pellets were resuspended in 300 µl PBS for FACS analysis or directly lysed for RNA and protein isolation.Exosomes from Caco-2 and HCT-116 CRC cells were analysed by: (1) Zetasizer Nano ZS (Malvern Instruments, UK); (2) flow cytometry for size determination and surface markers characterization, as previously reported [77]. Aldehyde/sulfate latex beads (Invitrogen, Sweden) (140,000) were incubated with 200 µl Caco-2 and HCT-116 exosomes at 37˚C for 30’ and then at 4˚C for 16 h on a rotator apparatus. After centrifugation at 4,000 g for 10’, pellets were resuspended in 100 µl PBS; 20 µl of 1 M glycine were added to block unspecific binding sites at 20°C for 30’. After one wash with PBS and 1% FBS, exosome-coated beads were incubated with PE-conjugated CD9, CD63 or CD81 antibodies or isotype controls (BD Biosciences) for 60’ at 4˚C. For FACS analysis, samples were washed and resuspended in 200 µl PBS/FBS and analysed with FACSCantoII (Becton Dickinson, San Diego CA) and FlowJo software (TreeStar). For cell viability assays, exosomes from treated or untreated source cells were isolated as described, resuspended in PBS and quantified by Qubit. 2 µg/100 µl or 5 µg/100 µl of exosomes were added to 9 x 103 recipient cells in 96 well plates and incubated for 24 h and 48 h. Cell viability was assessed through an MTT assay, 24 h and 48 h following exosome transfection. Absorbance values were read with a Multiscan Ascent microplate reader (Thermo Fisher Scientific). All MTT experiments were performed in six biological replicates. The statistical significance was evaluated by t-test (p-value < 0.05).
RNA isolation, reverse transcription and miRNA profiling by TaqMan Low Density Array
Total RNA was extracted with TriZol (Invitrogen), according to manufacturer instructions, and quantified by Qubit (Invitrogen). Samples (100 ng and 25 ng of total RNA from whole cell pellets or their exosomes, respectively) were retrotranscribed and preamplified [78]. Amplified products were loaded onto microfluidic cards of the TaqManHuman MicroRNA Array v3.0 A and B (Applied Biosystems). Experiments were performed in biological triplicates with a 7900HT Fast Real Time PCR System (Applied Biosystem). Result validation was obtained by single TaqMan assays (Applied Biosystems), according to the manufacturer’s instructions.
Analysis of miRNA expression data
To accurately profile miRNAs, we identified the most appropriate genes within the arrays that could be used for a reliable normalization of CRC cells or their exosomes. By applying two different methods, DataAssist v.3 software (Applied Biosystems) and the geNorm Algorithm [79], we pinpointed the following endogenous controls: miR-106a, miR-135b, miR-532-3p (Caco2 cells, panel A); miR-183*, miR-200a*, miR-1274B (Caco2 cells, panel B); miR-16, miR-138, miR-193b, (Caco2 exosomes, panel A); miR-9*, miR-183*, miR-766 (Caco2 exosomes, panel B); miR-185, miR-345, miR-362 (HCT-116 cells, panel A); miR-34a*, miR-1180, miR-1254 (HCT-116 cells, panel B); miR-106b, miR-135b, miR-301b (HCT-116 exosomes, panel A); miR-30a-3p, miR-148b*, miR-1274A (HCT-116 exosomes, panel B). MiRNAs expression changes were calculated by applying the 2-ΔΔCT method and using the above-mentioned miRNAs as endogenous controls. Differentially expressed (DE) miRNAs were identified by SAM (Significance of Microarrays Analysis) (http://www.tm4.org), applying a two-class paired test among ΔCt of treated and control samples (cells and exosomes) by using a p-value based on 100 permutations; imputation engine: K-nearest neighbours (10 neighbours); false discovery rate < 0.15. We accepted as reliable only DE miRNAs concordant by using all endogenous controls. Expression data in the Result section are shown as average relative quantity (RQ) of all values, calculated with each endogenous control respect to the calibrator sample. RQ values < 1 were converted to negative changes by following the formula: -1/RQ.
MiRNA targets analysis
Targets of DE miRNAs were identified by using a combination of different approaches: (i) structural predictions (http://mirecords.biolead.org); (ii) expression anticorrelation between miRNAs and their validated or putative mRNAs targets (http://mirgator.kobic.re.kr); (iii) data from Argonaute cross-linked immunoprecipitation-sequencing (CLIP-Seq) (http://starbase.sysu.edu.cn). The biological networks of DE miRNAs targets were built by retrieving the corresponding interactome data through NCBI Entrez Utilities Web Service Client. We determined the Gene Ontology (GO) functional classification of miRNA target networks through the tools DAVID (http://david.abcc.ncifcrf.gov) and FatiGO (http://babelomics3.bioinfo.cipf.es).
Antibody microarrays
To investigate the alterations of Caco-2 and HCT-116 exosomal protein profiles following Cetuximab treatment, proteins were extracted, labelled and incubated on antibody microarrays to detect abundance differences. This highly sensitive technology permits a proteomic analysis of small amounts of samples isolated from tissues, cultured cells or body liquids [80-82]. We used an array of 1,800 features, representing 810 antibodies, which are directed against 741 cancer-related proteins, and various controls [83] (Additional_file_1). Array production, quality control and handling were performed as described in much detail earlier [83, 84].
Exosomal protein extraction
Exosomal proteins were isolated from 28 different biological replicates (7 treated samples and 7 controls for both cell lines). Proteins were isolated by using an optimized lysis buffer [84]: 20% glycerol, 0.05 M bicine pH 8.5, 0.15 M NaCl, 0.002 M EDTA.2Na, 20 mM phenylmethanesulfonyl fluoride, 2% NP-40S, 1% sodium cholate, 0.25% n-dodecyl-β-D-maltoside (GenaXXon Bioscience, Ulm, Germany), 0.5% amidosulfobetaine-14, 1.0 U/μl of Benzonase (Merck Biosciences, Schwalbach, Germany), Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific, Bonn, Germany). Samples were incubated with 25 μl of lysis buffer for 1 h at 4°C on an orbital shaker, then they were centrifuged at 15,000 rpm at 4°C for 15’. The supernatant was aspirated with a fine needle, carefully avoiding to disturb the upper layer or the pellet. Protein concentration in the supernatant was determined with the Pierce BCA Protein Assay Kit using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific).
Protein labelling
Extracted protein samples were selected on the basis of their concentration; we studied only samples with a concentration of at least 1 mg/ml. If the value was higher, the protein concentration was adjusted to 1 mg/ml. Four biologic replicates of treated samples and five of controls were analyzed for both cell lines, making a total of 18 samples. For each sample, 20 µg protein were labelled as described in detail earlier [83, 84], by using the fluorescence dye DY-649 (Dyomics, Jena, Germany). A second fluorescence dye, DY-549, was used to label a pool of protein extracts from human serum samples of healthy people. The pool acts as a reference to normalize signal intensities in a dual-colour analysis mode [85]. Labelling reactions were performed at a molar ratio dye-protein of 7.5, with the assumption that 60 kDa is the average molecular weight of all proteins. Labelling was carried in the dark in 0.1 M carbonate buffer, pH 8.5, at 4°C for 2 h. Unreacted dye was quenched with 10% glycine for 20’ at 4°C on an orbital shaker in the dark. Labelled samples were stored at -20° C until analysis.
Sample incubation
Incubation of the microarrays with labelled samples was performed as reported [84]. In short, the antibody arrays were soaked in TBST buffer. Subsequently, samples were washed once for 5’ with TBST containing 0.05% Tween-20 (TBST20) (pH 6.5), followed by another wash for 15’ with TBST20 (pH 7.5). Slides were blocked in 5.0 ml of 10% non-fat dry milk (Biorad, Munich, Germany), at 20°C for 3 h, using Quadriperm chambers (Greiner Bio-One, Frickenhausen, Germany) on an orbital shaker. Blocked slides were incubated in Quadriperm chambers with 20 μg of DY-649 labelled sample and 20 μg of DY-549 labelled pool reference in 4 ml incubation buffer, containing 1% milk in PBST20 (pH 7.5), on an orbital shaker in the dark at 4°C for 16 h. Slides were washed four times, 5’ each, in large volumes of PBST20 (pH 7.5), rinsed with deionised water for 10’, and dried in a ventilated oven at 29°C. Scanning of the arrays was performed with a ScanArray-4000XL (Perkin Elmer,Waltham, USA) at constant laser power and photomultiplier tube gain (PMT). Images were analysed with the software GenePix Pro 6.0 (Molecular Devices, Sunnyvale, USA).
Antibody microarray data analysis
GPR files of scanned images were analysed with Chipster software (v1.4.6, CSC, Finland). Data were normalized using normexp with background correction offset [0,50], as reported [86]. Similarities and differences among different sample groups were globally assessed using hierarchical clustering, non-metric multidimensional scaling [87], and detrended correspondence analysis [88]. Two-group comparisons (ie, Caco-2 cells versus HCT-116 cells, both for treated and control exosome samples; treated samples versus controls from the same cell line) were performed using the Empirical Bayes test with Bonferroni–Hochberg multiple testing correction; cut-off was set at a p-value of 0.05 [89]. Multiple-group comparisons (ie, for cell of origin and degree of differentiation) were performed using LIMMA with a p-value adjustment according to Bonferroni–Hochberg multiple testing correction [89]. Cluster analysis was conducted using Pearson correlations; dendrograms were constructed using the average linkage method (http://chipster.csc.fi). Functional investigations were performed with the Ingenuity Systems Pathway Analysis tools (Ingenuity Systems, Redwood City, CA). The p-values were calculated using right-tailed Fisher’s exact test (http://www.ingenuity.com).
ACKNOWLEDGEMENTS
We gratefully acknowledge the kind gift of CACO-2 and HCT-116 cells by Prof Massimo Romani (IST, Genova, Italy). Cetuximab was kindly provided by Dr Dario Giuffrida (Istituto Oncologico del Mediterraneo, Catania, Italy). This project was funded by MIUR (MP, CDP, AC, GLD) and the European Commission as part of the Affinomics consortium as well as the MOST-DKFZ initiative (JDH, MSSA). We also would like to acknowledge EMBO and Wood-Whelan research fellowship for supporting LS and MM, respectively, with short-term fellowships.


- 1. Get on the exosome bus with ALIX. Nat Cell Biol. 2012 Jun 29; 14(7):654-5. https://doi.org/10.1038/ncb2530. [PubMed].
- 2. Urbanelli L, Magini A, Buratta S, Brozzi A, Sagini K, Polchi A, Tancini B, Emiliani C. Signaling Pathways in Exosomes Biogenesis, Secretion and Fate. Genes 2013; 4(2):152-170. https://doi.org/10.3390/genes4020152. [PubMed].
- 3. Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods 2002; 270:211–226. https://doi.org/10.1016/s0022-1759(02)00330-7. [PubMed].
- 4. Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem 1998; 273:20121–20127. https://doi.org/10.1074/jbc.273.32.20121. [PubMed].
- 5. Kapsogeorgou EK, Abu-Helu RF, Moutsopoulos HM, Manoussakis MN. Salivary gland epithelial cell exosomes: A source of autoantigenic ribonucleoproteins. Arthritis Rheum 2005; 52:1517–1521. https://doi.org/10.1002/art.21005. [PubMed].
- 6. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. 2011; 2011:842849. https://doi.org/10.1155/2011/842849. [PubMed].
- 7. Proteomic Analysis of Human Parotid Gland Exosomes by Multidimensional Protein Identification Technology (MudPIT). J Proteome Res. 2009 Mar; 8(3):1304-14. https://doi.org/10.1021/pr800658c. [PubMed].
- 8. Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched. RNA. 2008; 14:1424–1432. https://doi.org/10.1261/rna.755908. [PubMed].
- 9. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007; 72:1095–1102. https://doi.org/10.1038/sj.ki.5002486. [PubMed].
- 10. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol Oncol. 2007; 107:563–571. https://doi.org/10.1016/j.ygyno.2007.08.064. [PubMed].
- 11. Exosomes from bronchoalveolar fluid of tolerized mice prevent allergic reaction. J Immunol. 2008; 181:1519–1525. https://doi.org/10.4049/jimmunol.181.2.1519. [PubMed].
- 12. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007; 179:1969–1978. https://doi.org/10.4049/jimmunol.179.3.1969. [PubMed].
- 13. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004 Nov 15; 104(10):3257-66. https://doi.org/10.1182/blood-2004-03-0824. [PubMed].
- 14. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009 Aug; 9(8):581-93. https://doi.org/10.1038/nri2567. [PubMed].
- 15. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007 Jun; 9(6):654-9. https://doi.org/10.1038/ncb1596. [PubMed].
- 16. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011 May 15; 81(10):1171-82. https://doi.org/10.1016/j.bcp.2011.02.011. [PubMed].
- 17. The Jeanne Manery-Fisher Memorial Lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochem Cell Biol. 1992 Mar-Apr; 70(3-4):179-90. https://doi.org/10.1139/o92-028. [PubMed].
- 18. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009 Jun; 6(3):267-83. https://doi.org/10.1586/epr.09.17. [PubMed].
- 19. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011 Feb; 11(4):709-20. https://doi.org/10.1002/pmic.201000422. [PubMed].
- 20. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004; 5:317–323. https://doi.org/10.1038/nrm1360. [PubMed].
- 21. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013 Dec 20; 4:2980. https://doi.org/10.1038/ncomms3980. [PubMed].
- 22. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One. 2010 Dec 17; 5(12):e15353. https://doi.org/10.1371/journal.pone.0015353. [PubMed].
- 23. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010 Sep 10; 73(10):1907-20. https://doi.org/10.1016/j.jprot.2010.06.006. [PubMed].
- 24. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009 Feb 26; 113(9):1977-81. https://doi.org/10.1182/blood-2008-08-174094. [PubMed].
- 25. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004 Jun 29; 101(26):9683-8. https://doi.org/10.1073/pnas.0308413101. [PubMed].
- 26. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. 2012 Aug; 22(4):342-9. https://doi.org/10.1016/j.semcancer.2012.02.005. [PubMed].
- 27. Record, M. Exosomal Lipids in Cell–Cell Communication. In Emerging Concepts of Tumor Exosome–Mediated Cell-Cell Communication. Zhang, H.-G., Ed.; Springer: New York, NY, USA. 2013; pp. 47–68.
- 28. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008 Jul; 110(1):13-21. https://doi.org/10.1016/j.ygyno.2008.04.033. [PubMed].
- 29. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012 Apr; 51(4):409-18. https://doi.org/10.1002/gcc.21926. [PubMed].
- 30. Targeted therapy in metastatic colorectal cancer -- an example of personalised medicine in action. Cancer Treat Rev. 2013 Oct; 39(6):592-601. https://doi.org/10.1016/j.ctrv.2012.12.011. [PubMed].
- 31. Individualized therapies in colorectal cancer: KRAS as a marker for response to EGFR-targeted therapy. J Hematol Oncol. 2009 Apr 22; 2:18. https://doi.org/10.1186/1756-8722-2-18. [PubMed].
- 32. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol Cancer Ther. 2010 Dec; 9(12):3396-409. https://doi.org/10.1158/1535-7163.MCT-10-0137. [PubMed].
- 33. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012 Feb; 227(2):658-67. https://doi.org/10.1002/jcp.22773. [PubMed].
- 34. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996 Mar 1; 183(3):1161-72. https://doi.org/10.1084/jem.183.3.1161. [PubMed].
- 35. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013 Feb 18; 200(4):373-83. https://doi.org/10.1083/jcb.201211138. [PubMed].
- 36. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011 Dec; 12(12):1659-68. https://doi.org/10.1111/j.1600-0854.2011.01225.x. [PubMed].
- 37. Exosomes: Fit to deliver small RNA. Commun Integr Biol. 2010 Sep; 3(5):447-50. https://doi.org/10.4161/cib.3.5.12339. [PubMed].
- 38. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005 Oct; 4(10):1595-604. https://doi.org/10.1158/1535-7163.MCT-05-0102. [PubMed].
- 39. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011 Dec; 46(12):1391-402. https://doi.org/10.1007/s00535-011-0456-0. [PubMed].
- 40. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol. 2011 May; 48(9-10):1084-90. https://doi.org/10.1016/j.molimm.2011.02.001. [PubMed].
- 41. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012 Oct; 61(10):1447-53. https://doi.org/10.1136/gutjnl-2011-301122. [PubMed].
- 42. Role of miR-150-targeting c-Myb in colonic epithelial disruption during dextran sulphate sodium-induced murine experimental colitis and human ulcerative colitis. J Pathol. 2011 Dec; 225(4):544-53. https://doi.org/10.1002/path.2907. [PubMed].
- 43. Dynamic nature of noncoding RNA regulation of adaptive immune response. Int J Mol Sci. 2013 Aug 22; 14(9):17347-77. https://doi.org/10.3390/ijms140917347. [PubMed].
- 44. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007 Apr 24; 104(17):7080-5. https://doi.org/10.1073/pnas.0702409104. [PubMed].
- 45. MicroRNA-433 negatively regulates the expression of thymidylate synthase (TYMS) responsible for 5-fluorouracil sensitivity in HeLa cells. BMC Cancer. 2013 Aug 2; 13(1):369. https://doi.org/10.1186/1471-2407-13-369. [PubMed].
- 46. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res. 2009 Jun 16; 28:82. https://doi.org/10.1186/1756-9966-28-82. [PubMed].
- 47. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol. 2010 Sep; 11(9):806-13. https://doi.org/10.1038/ni.1916. [PubMed].
- 48. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol. 2011 Oct; 22(10):2257-66. https://doi.org/10.1093/annonc/mdq758. [PubMed].
- 49. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. J Immunol. 2011 Dec 1; 187(11):5834-41. https://doi.org/10.4049/jimmunol.1100922. [PubMed].
- 50. Ding S, Liang Y, Zhao M, Liang G, Long H, Zhao S, Wang Y, Yin H, Zhang P, Zhang Q, Lu Q. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 2012 Sep; 64(9):2953-63. https://doi.org/10.1002/art.34505. [PubMed].
- 51. Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB J. 2008 Sep; 22(9):3358-69. https://doi.org/10.1096/fj.07-102855. [PubMed].
- 52. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002 May 20; 195(10):1303-16. https://doi.org/10.1084/jem.20011624. [PubMed].
- 53. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005 Jun; 128(7):1796-804. https://doi.org/10.1053/j.gastro.2005.03.045. [PubMed].
- 54. Tumour exosomes inhibit binding of tumour-reactive antibodies to tumour cells and reduce ADCC. Cancer Immunol Immunother. 2011 May; 60(5):639-48. https://doi.org/10.1007/s00262-011-0979-5. [PubMed].
- 55. Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life Sci. 2011 Aug 29; 89(9-10):304-12. https://doi.org/10.1016/j.lfs.2011.06.020. [PubMed].
- 56. Cancer cells induced to express mesenchymal phenotype release exosome-like extracellular vesicles carrying tissue factor. J Biol Chem. 2012 Dec 21; 287(52):43565-72. https://doi.org/10.1074/jbc.M112.401760. [PubMed].
- 57. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. 2013 Jun; 31(5):330-5. https://doi.org/10.3109/07357907.2013.789905. [PubMed].
- 58. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010 Sep 24; 285(39):30139-49. https://doi.org/10.1074/jbc.L110.145698. [PubMed].
- 59. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012 Aug 1; 122(8):2884-97. https://doi.org/10.1172/JCI63455. [PubMed].
- 60. Expression of miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in αβ and γδ T cells. PLoS One. 2011; 6(5):e20171. https://doi.org/10.1371/journal.pone.0020171. [PubMed].
- 61. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res. 2013 Oct 1; 100(1):7-18. https://doi.org/10.1093/cvr/cvt161. [PubMed].
- 62. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J Biol Chem. 2011 Jul 29; 286(30):26487-95. https://doi.org/10.1074/jbc.M110.213561. [PubMed].
- 63. CD59 blockade enhances antigen-specific CD4+ T cell responses in humans: a new target for cancer immunotherapy?. J Immunol. 2009 May 1; 182(9):5203-7. https://doi.org/10.4049/jimmunol.0804243. [PubMed].
- 64. A network analysis of the human T-cell activation gene network identifies JAGGED1 as a therapeutic target for autoimmune diseases. PLoS One. 2007 Nov 21; 2(11):e1222. https://doi.org/10.1371/journal.pone.0001222. [PubMed].
- 65. AGR2 is a novel surface antigen that promotes the dissemination of pancreatic cancer cells through regulation of cathepsins B and D. Cancer Res. 2011 Nov 15; 71(22):7091-102. https://doi.org/10.1158/0008-5472.CAN-11-1367. [PubMed].
- 66. TRIM22: A Diverse and Dynamic Antiviral Protein. Mol Biol Int. 2012; 2012:153415. https://doi.org/10.1155/2012/153415. [PubMed].
- 67. Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J Immunol. 2012 Jun 15; 188(12):5954-61. https://doi.org/10.4049/jimmunol.1103466. [PubMed].
- 68. Molecular response to cetuximab and efficacy of preoperative cetuximab-based chemoradiation in rectal cancer. J Clin Oncol. 2009 Jun 10; 27(17):2751-7. https://doi.org/10.1200/JCO.2008.18.5033. [PubMed].
- 69. Inflammation and cancer. Nature. 2002 Dec 19-26; 420(6917):860-7. https://doi.org/10.1038/nature01322. [PubMed].
- 70. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009 Jul; 30(7):1073-81. https://doi.org/10.1093/carcin/bgp127. [PubMed].
- 71. The origins of tumor-promoting inflammation. Cancer Cell. 2013 Aug 12; 24(2):143-4. https://doi.org/10.1016/j.ccr.2013.07.016. [PubMed].
- 72. Molecular epidemiology, cancer-related symptoms, and cytokines pathway. Lancet Oncol. 2008; 9(8):777-85. https://doi.org/10.1016/S1470-2045(08)70197-9. [PubMed].
- 73. Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol Oncol. 2012 Dec; 6(6):611-9. https://doi.org/10.1016/j.molonc.2012.09.005. [PubMed].
- 74. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med. 2012 Jun 27; 10:134. https://doi.org/10.1186/1479-5876-10-134. [PubMed].
- 75. Evolutionary implications of horizontal gene transfer. Annu Rev Genet. 2012; 46:341-58. https://doi.org/10.1146/annurev-genet-110711-155529. [PubMed].
- 76. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012 Oct 15; 21(R1):R125-34. https://doi.org/10.1093/hmg/dds317. [PubMed].
- 77. Wahlgren J, De L Karlson T, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H: Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012 Sep 1; 40(17):e130. https://doi.org/10.1093/nar/gks463. [PubMed].
- 78. Specific alterations of the microRNA transcriptome and global network structure in colorectal cancer after treatment with MAPK/ERK inhibitors. J Mol Med (Berl). 2012 Dec; 90(12):1421-38. https://doi.org/10.1007/s00109-012-0918-8. [PubMed].
- 79. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002 Jun 18; 3(7):RESEARCH0034. https://doi.org/10.1186/gb-2002-3-7-research0034. [PubMed].
- 80. Affinity-based microarrays for proteomic analysis of cancer tissues. Proteomics Clin Appl. 2013; 7: 8-16. https://doi.org/10.1002/prca.201200114. [PubMed].
- 81. Immunoassay-based proteome profiling of 24 pancreatic cancer cell lines. J Proteomics. 2012; 75: 3747-3759. https://doi.org/10.1016/j.jprot.2012.04.042. [PubMed].
- 82. Schröder C, Srinivasan H, Sill M, Linseisen J, Fellenberg K, Becker N, Nieters A, Hoheisel, JD. Plasma protein analysis of patients with different B-cell lymphomas with high-content antibody microarrays. Proteomics Clin Appl. 2013; 7: 802-812. https://doi.org/10.1002/prca.201300048. [PubMed].
- 83. Dual-color proteomic profiling of complex samples with a microarray of 810 cancer-related antibodies. Mol Cell Proteomics. 2010 Jun; 9(6):1271-80. https://doi.org/10.1074/mcp.M900419-MCP200. [PubMed].
- 84. Analysis conditions for proteomic profiling of mammalian tissue and cell extracts with antibody microarrays. Proteomics. 2010 Sep; 10(17):3203-7. https://doi.org/10.1002/pmic.201000170. [PubMed].
- 85. Assessment and optimisation of normalisation methods for dual-colour antibody microarrays. BMC Bioinformatics. 2010; 11: 556. https://doi.org/10.1186/1471-2105-11-556. [PubMed].
- 86. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007 Oct 15; 23(20):2700-7. https://doi.org/10.1093/bioinformatics/btm412. [PubMed].
- 87. Relational patterns of gene expression via non-metric multidimensional scaling analysis. Bioinformatics. 2005 Mar; 21(6):730-40. https://doi.org/10.1093/bioinformatics/bti067. [PubMed].
- 88. Correspondence analysis applied to microarray data. Proc Natl Acad Sci U S A. 2001 Sep 11; 98(19):10781-6. https://doi.org/10.1073/pnas.181597298. [PubMed].
- 89. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004; 3:Article3. https://doi.org/10.2202/1544-6115.1027. [PubMed].
Last Modified: 2016-06-14 10:11:35 EDT
