Oncoscience
HER2 and uPAR cooperativity contribute to metastatic phenotype of HER2-positive breast cancer
Vineesh Indira Chandran1, Serenella Eppenberger-Castori2, Thejaswini Venkatesh3, Kara Lea Vine4,5,6, Marie Ranson4,5,6
1 Department of Clinical Sciences, Section of Oncology and Pathology, Lund University, Lund, Sweden.
2 Institute for Pathology, Department of Molecular Pathology, Schoenbeinstrasse, Basel, Switzerland.
3 Nitte University Centre for Science Education and Research (NUCSER), K. S. Hegde Medical Academy, NitteUniversity, Deralakatte, Mangalore, Karnataka, India.
4 School of Biological Sciences, University of Wollongong, Wollongong, NSW, Australia.
5 Centre for Medical & Molecular Bioscience, University of Wollongong, Wollongong, NSW, Australia.
6 Illawarra Health and Medical Research Institute, University of Wollongong, Wollongong, NSW, Australia.
Correspondence to: Vineesh Indira Chandran, email: [email protected]
Keywords: HER2/ERBB2, uPAR/PLAUR, HER2-positive breast cancer, co-overexpression, co-amplification, correlation
Received: January 20, 2015
Accepted: March 16, 2015
Published: March 23, 2015
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
Human epidermal growth factor receptor type 2 (HER2)-positive breast carcinoma is highly aggressive and mostly metastatic in nature though curable/manageable in part by molecular targeted therapy. Recent evidence suggests a subtype of cells within HER2-positive breast tumors that concomitantly expresses the urokinase plasminogen activator receptor (uPAR) with inherent stem cell/mesenchymal-like properties promoting tumor cell motility and a metastatic phenotype. This HER-positive/uPAR-positive subtype may be partially responsible for the failure of HER2-targeted treatment strategies. Herein we discuss and substantiate the cumulative preclinical and clinical evidence on HER2-uPAR cooperativity in terms of gene co-amplification and/or mRNA/protein co-overexpression. We then propose a regulatory signaling model that we hypothesize to maintain upregulation and cooperativity between HER2 and uPAR in aggressive breast cancer. An improved understanding of the HER2/uPAR interaction in breast cancer will provide critical biomolecular information that may help better predict disease course and response to therapy.
INTRODUCTION
Breast cancer (BC) is a highly heterogeneous disease consisting of several subtypes, each classified by their unique biological signature [1, 2]. Each BC subtype exhibits varied responses to different therapeutic regimens. Treatment options for metastatic disease remains limited despite the availability of several United States Food and Drug Administration (FDA) approved drugs against BC [3]. In this scenario, it is imperative to explore different therapeutic models of targeting one or more tumor-specific biomarkers that define the more aggressive breast carcinoma subtypes efficiently for improved management of the disease.Established BC biomarkers predicting metastatic risk include lymph-node involvement, hormone independency, loss of histopathological differentiation of primary tumor (grade), elevated proliferation, and angiogenesis. However, these biomarkers confidently predict outcome for only ~30% of patients. Of the remaining patients some will still develop metastases whilst others will not [4]. Components of the urokinase plasminogen activation system, particularly urokinase plasminogen activator (uPA, Gene symbol: PLAU; located on chromosome 10q22.2), its receptor uPAR (Gene symbol: PLAUR; located on chromosome 19q13) and inhibitor plasminogen activator inhibitor type 1 (PAI-1, Gene symbol: SERPINE1; located on chromosome 7q22.1) are proven to be associated with aggressive carcinoma. The combination of uPA/PAI-1 at the protein level is a strong and independent predictor of metastasis in lymph-node negative BC patients and predicts response to hormone therapy [5, 6]. uPAR is expressed in malignant cells and in the tumor stroma which translates into an aggressive tumor phenotype and poor relapse-free survival (RFS) [7].
The recognition of human epidermal growth factor receptor type 2 (HER2, Gene Symbol HER2; located on chromosome 17q12) over-expression as a therapeutic target for advanced breast carcinoma was primarily related to the clinical finding that HER2/neu proto-oncogene is amplified in 15–25% of all breast tumors, and is often associated with poor disease-free survival (DFS) [8-15]. The mechanism by which HER2 overexpression imparts increased aggressiveness to tumors has been attributed mostly to dysregulated activation of downstream intracellular signaling pathways [16-25]. In some cases HER2 overexpression has been reported to induce resistance to certain chemotherapeutics [26-28]. Furthermore, HER2 overexpression has been found in both in the primary tumor, circulating tumor cells (CTCs) and corresponding metastases [29-31].
A high level of correlation was observed between HER2 and uPAR mRNA in disseminated tumor cells (DTCs) in 8 out of 16 patients (50%) and was associated with a more aggressive primary tumor phenotype (estrogen receptor (ER)-negative, progesterone receptor (PR)-negative or HER2-positive) [32]. Also a positive association between HER2 and PLAUR gene amplification (which was concordant with protein expression in both cases) was found in >90% of HER2-amplified individual tumor cells from the blood or tissue of patients with advanced recurrent BC [33]. These and other studies [34-38] suggested the possibility of cooperativity between the HER2 and uPAR signaling pathways leading to recurrence/metastases; however the exact mechanism remains to be elucidated. Furthermore, nuclear factor-kappaB (NF-κB) mediated expression of HER2 and uPAR in cancer stem cells (CSCs), has been implicated for maintaining malignancy at the invasive edge of BC, which suggests an enhanced role for HER2-uPAR cooperative overexpression in disease relapse with an aggressive intent [39].This review analyzes and substantiates the cooperativity between HER2 and PLAUR in terms of their correlation status at the mRNA level in primary tumors of BC patients. For the first time, we also propose a regulatory signaling model as a mechanism responsible for maintaining the aggressive properties of primary and DTCs, through high co-expression of HER2 and uPA receptors and use it as a rationale to highlight the importance of simultaneously targeting HER2 and uPAR in advanced BC.
HER2-positive BC
A working model for BC molecular taxonomy utilizing microarray-based gene expression profiling classifies BCs by hierarchical cluster analysis, using an intrinsic gene list, into four main molecular subtypes: luminal A, luminal B, basal-like, and HER2 [40-45], with subgroups increasingly being identified such as claudin-low and normal breast-like [46-49]. Each subtype displays unique patterns of metastatic spread associated with notable differences in survival after relapse [50]. Clinically, HER2-positive tumors comprise approximately 12–30% of all invasive BCs and are most often found in younger patients and associated with poorer clinical outcomes [51, 52]. This subtype is associated with increased cell proliferation, angiogenesis, tumor invasiveness, and a high nuclear grade [53]. It has been observed that patients with HER2-positive tumors are more likely to have multifocal/multicentric cancers and nodal involvement [54]. At the molecular level, HER2-positive BCs exhibits extensive changes in the patterns of gene expression associated with the HER2 pathway and/or HER2 amplicon located in the 17q12 chromosome. The manifestation of the variation in the expression of specific subsets of genes exclusive to HER2-positive BC is reflected mainly in the variation in growth rate, activity of specific signaling pathways, and in the cellular composition of the tumors [40]. Several signaling pathways are triggered in HER2-positive BC [55-57]. A detailed description of HER2-positive BC subtype can be found in Eroles et al. [49].
uPAR expression in BC
The urokinase receptor (uPAR) is linked to the plasma membrane via a glycosyl phosphatidylinositol (GPI) anchor, which is hypothesized to enable high intramembrane mobility [58]. Upon binding uPA with high affinity (1 nM) and selectivity, co-localized zymogen plasminogen is converted to the serine proteinase plasmin thereby facilitating cell migration by tissue remodeling. uPAR interacts with other molecules disparate from its function as a proteinase receptor, including vitronectin, members of the integrin adhesion receptor superfamily, caveolin, and G-protein-coupled receptor (GPCR). As a result, uPAR activates intracellular signaling molecules such as tyrosine- and serine-protein kinases (such as EGF receptor, lymphocyte protein tyrosine kinase (Lck), haematopoietic cell kinase (Hck), Src, focal adhesion kinase (FAK) and extracellular-signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK)) ultimately affecting migration, adhesion, differentiation and proliferation through intracellular signaling [59, 60]. Numerous clinical studies have implicated uPAR expression with phenotypically aggressive BC [61, 62] and low DFS [63]. Tumor cells (e.g. MCF-7, LNCaP) that do not express uPAR or express only low levels of uPAR were poorly tumorigenic in mice [64]. uPAR expression has often been found restricted to cells at the invasive edge of a tumor or in tumor cells at the tumor-stromal interface [65] and the expression of uPAR appears to increase with grade or stage of the tumor and may be enriched in metastatic lesions [66]. Various studies have also found uPAR to be highly expressed in CTCs from patients with advanced breast cancers [33, 38]. In addition, uPAR expression has been described in CSCs in BC [67]. For example, Jo et al., [67] showed that MCF-7 and MDA-MB-468 BC cells acquire CSC-like properties when uPAR is overexpressed and uPAR-dependent signaling is activated.
HER2 and uPAR cooperativity in HER2-positive BC
Cooperation of HER2 and uPAR at mRNA level
Cooperativity between HER2 and uPAR has emerged as a strong determinant for the aggressive properties of HER2-positive BC [33, 34, 37]. Although HER2 and uPAR were described as independent tumor-specific protein predictors of BC progression for decades, correlative expression of HER2 and uPAR was first reported by Pierga et al., [32] who found a high level of correlation between HER2 and uPAR mRNA in disseminated tumor cells (DTC) in 8 out of 16 patients (50%) and was associated with a more aggressive primary tumor phenotype (estrogen receptor (ER)-negative, progesterone receptor (PR)-negative or HER2-positive). Following this, Meng et al., [33] found PLAUR to be co-amplified with HER2 in individual tumor cells in the blood and tissue of advanced recurrent primary BC patients. They found that if the advanced BC patients had higher HER2 gene amplification in tumor cells from their primary breast carcinomas, then they were more likely to have co-amplification and higher levels of PLAUR amplification. They observed 92% (23 of 25) PLAUR gene amplification in HER2 amplified cases in touch preps of primary tumor and CTCs; whereas in HER2 nonamplified tumors, only 3% (1 of 39) were PLAUR gene amplified, highlighting the correlation of HER2 and PLAUR gene status.
Similarly, another independent study published by Urban et al., [35], in the same year, showed that patients with HER2-positive/PLAU–positive tumors (as assessed at the mRNA level) exhibited significantly reduced metastases-free survival (MFS) compared to patients with HER2-positive/PLAU–negative tumors. This study strongly implicated uPA expression, using three independent study populations assayed by different gene expression techniques, as a powerful prognostic indicator associated with distant MFS in patients with HER2-positive tumors. This was later confirmed by Staaf et al., [36] who found PLAU gene status within the HER2-derived prognostic predictor (HDPP) gene signature strongly associated with basal-like, ER-negative, lymph-node positive, high grade BC. They found a significant correlation between increased mRNA and protein levels of the PLAU gene in tumors classified as poor by HDPP and in the data set obtained from the Nederlands Kanker Instituut (NKI), HDPP retained strong prognostic value when stratified for PLAU status for both overall survival (OS) and distant metastasis-free survival (DMFS). Very recently, Berg et al., [37] found significant correlation between HER2 and uPAR while analyzing protein networks in 106 formalin-fixed and paraffin-embedded (FFPE) BC tissues by reverse phase protein microarray (RPPA) analysis. Markiewicz et al., [38] found that CTC-enriched HER2-positive (mRNA) blood samples from lymph node positive BC patients were 100% positive for PLAUR mRNA expression compared to 34% of HER2 negative samples. Further, in the hierarchical clustering of the clinicopathological data, where the study population was divided into two main groups that differed in the expression of VIM, CXCR4, PLAUR, HER2, they found that patients in the cluster with elevated expression of these genes showed more frequent lymph node involvement (58%) than patients from the cluster with lower expression (35%). All this evidence point towards a potential strong cooperativity between HER2 and uPAR resulting in enhanced metastatic potential, giving early indications of a potential synergistic co-expression.
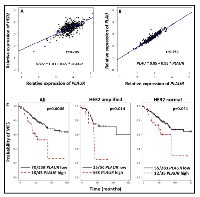
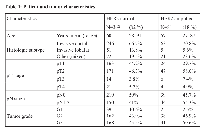
In order to ascertain the interdependence between HER2 and PLAUR mRNA status in HER2-positive breast carcinoma, we constructed a correlation curve utilizing the HER2 and PLAUR mRNA values obtained from fresh frozen tissue of the former “Stiftung Tumorbank Basel” biobank (now part of the Biobank Pathology at the University Hospital of Basel, BPUB). In this subset of 450 primary BC patients, we found the RNA expression levels of HER2 and PLAUR to be strongly and significantly (r=0.705, p<0.0001) correlated (Figure 1A). An extremely strong correlation (r= 0.954, p<0.0001) between the gene expression levels of PLAU and its receptor PLAUR was also observed (Figure 1B). Patients and tumor characteristics are displayed in Table 1.
Furthermore, we performed Kaplan-Meier analyses with respect to MFS in the overall collective as well as in the subset with normal HER2 (82%) and amplified HER2 (18%). Figure 1C illustrates the strong impact of PLAUR overexpression in the overall collective and the two HER2 subsets. PLAUR overexpression correlated with poor outcome in the overall cohort of patients. Of interest PLAUR retained a significant impact also in the subsets with HER2 amplification (See Figure 1C, HER2 amplified). In the overall collective, the probability of MFS at 5 years for patients with PLAUR overexpressing tumors was 0.524 (CI: 0.373- 0.735) as compared to 0.770 (CI: 0.718-0.826) for those with low PLAUR expression levels. These values decreased to 0.250 (CI: 0.075-0.830) and 0.716 (CI: 0.598-0.858) in the HER2 amplified subset. Moreover, the Kaplan-Meier curves depicted better MFS for patients with HER2 normal and low PLAUR phenotype tumors. In this case the following rates at five years were calculated: 0.601 (CI: 0.427- 0.845) for high and 0.781 (CI: 0.723-0.843) for low PLAUR expression levels, respectively.
However, unlike the high and strong correlation between HER2 and PLAUR mRNA expression, the correlation in the HER2 and PLAUR gene co-amplification status in primary BC patients analyzed has been infrequent or absent. This is not surprising and is consistent with previous reports where HER2 and PLAUR co-amplification status has been found to be a rare event across primary BC patients [68]. This is further evident in The Cancer Genome Atlas (TCGA) Breast Invasive Carcinoma Project data which involved analysis of primary BC s by genomic DNA copy number arrays, DNA methylation, exome sequencing, messenger RNA arrays, microRNA sequencing and reverse-phase protein arrays. This study found only one case of co-amplification of HER2 and PLAUR in 825 primary BC patients [69, 70]. It should also be noted that by analyzing individual tumor cells the effect of averaging out gene amplification and/or expression status in tumors or their metastases is negated [33]. That is, significant associations between HER2 and PLAUR gene co-amplification and co-expression may not be seen when biopsies of mixed cell populations are analyzed.Common signaling molecules downstream of HER2 and uPAR
The hypothesis that high uPAR expression could be required for the invasive capacity of HER2 positive tumors was demonstrated by Tan et al., [34] who showed that uPA system contributes to a higher metastatic potential in HER2-overexpressing cancer cells. In HER2-overexpressing BC cells, Tan et al., [34] found upregulation and activation of protein kinase Cα (PKCα) through steroid receptor co-activator (Src) by HER2 to be critical for HER2-mediated cancer cell invasion. Other studies have found PKCα and Src to be critical components for uPAR-mediated cancer cell invasion in high uPAR expressing cancer cells [71, 72]. Tan et al. [34] also found that by inhibiting PKCα or Src by chemical inhibitors, dominant-negative mutants or siRNA, uPAR expression decreased and there was a reduction in cancer cell invasion in HER2 overexpressing BC cell lines. This indicates that HER2-mediated PKCα/Src upregulation and activation is required for the HER2-mediated upregulation of the uPAR, which may contribute to invasion and metastasis in HER2 positive tumors.Src downstream of HER2 and uPAR
The p160 Src family contains 3 members: Src-1 (nuclear receptor co-activator 1 (NCOA1)), Src-2 (transcriptional intermediary factor-2 (TIF2), glucocorticoid receptor interacting protein-1 (GRIP1), or NCOA2), and Src-3 (amplified in BC-1 (AIB1), activator of retinoid and thyroid receptors (ACTR), or NCOA3) [73]. The SRC family members share an overall similarity of 50–55% in their amino acid sequences and interact with and coactivate other transcription factors such as ETS-2, PEA3, and E2F1 [74-83]. Numerous studies have been reported that show Src as a proto-oncoprotein of BC. Src binds to HER2 and is activated in HER2-overexpressing cancer cells [34, 84-86]. Among the Src family members, high Src-1 expression has been directly correlated with HER2 positivity, disease recurrence in HER2-positive BCs and resistance to endocrine therapy [77, 78], and disruption of the Src-1 gene in mice suppresses BC metastasis without affecting primary tumor formation [82]. Many other studies have also positively correlated Src with HER2 positive BC [87]. Meanwhile, Src has also already been shown to transduce signals from uPAR [88] providing mammary MCF-7 cells with a proliferative and invasive advantage.HER2 and uPAR signaling mediated by PKCα
PKC family comprises of several isoforms that belongs to the family of serine/threonine kinases that regulate cell proliferation, differentiation, apoptosis, motility and adhesion [89, 90]. Various studies have found the PKC isoforms, mainly PKCα, PKCδ, and PKCε, to be highly expressed in BC cells imparting them with an increased invasive or metastatic potential than in normal tissues [34, 91-95]. For a detailed overview of the role of each of the PKC isoforms on tumorigenesis and BC in particular, refer to Lønne et al., [96]. Early experimentations by Peles et al., [97] showed HER2 to activate PKCα via phospholipase-γ (PLC γ). However, direct evidence on the exact regulatory role of PKCα expression in BC downstream of HER2 only came to known following studies by Tan et al., [34] as mentioned previously. Recently, Magnifico et al., [98] showed a specific physical association between PKCα and HER2 using solubilized lipid rafts and demonstrated PKCα mediated upregulation of HER2 expression and vice versa. They found, in HER2 positive BC cells, PKCα inhibition by pharmacologic treatments and PKCα-specific small interfering RNA (siRNA) led to a dramatic downregulation of HER2 levels. Consistent with this inhibition of HER2 activation by the tyrosine kinase inhibitor lapatinib led to decreased levels of PKCα phosphorylation. Thus PKCα has come to be recognized as a potential marker for BC aggressiveness. More importantly, Magnifico et al., [98] showed that HER2 overexpression in HER2 positive carcinomas is predominantly regulated by PKCα activity. The manifestation of this finding could be a regulatory loop where high PKCα expression maintains the HER2 overexpression and hence invasiveness. With respect to uPAR, studies reported as early as 1994 by Busso et al., [99] found that uPAR forms complexes with PKC in epithelial cells. Further to this, Sliva et al., [100] showed that inhibition of PKC represses constitutive (nonstimulated) migration of highly metastatic MDA-MB-231 cells with constitutively high levels of uPA. In this scenario, similar to PKC regulation of HER2 overexpression as mentioned earlier, it can be assumed that the constitutive uPA expression is maintained non-canonically by activation of PKC.NF-κB pathway intermediates signaling from HER2 and uPAR
Both HER2 and uPAR are interlinked to NF-κB signaling. HER2 activates NF-κB signaling in HER2 overexpressing BC cell lines [101]. The canonical NF-κB family pathway, that is overexpressed in BC cells from both primary human tumors and in cell lines [102], mediates HER2-induced breast CSC expansion [103]. This finding implicating HER2 expression with CSC expansion is supported by several lines of evidence from independent studies. In one such study, the overexpression of HER2 correlated with the expression of the stem cell marker aldehyde dehydrogenase (ALDH) in BC patients [104]. Cicalese et al., [105] found that increased HER2 transgene expression in mice resulted in increased self-renewal and replicative potential for CSCs. In a separate in vitro study on BC cells, Korkaya et al., [106] found that HER2 overexpression increased the CSC population, as demonstrated by increased ALDH activity, mammosphere formation, tumorigenesis, and expression of stem cell related genes. On the other hand, NF-κB and other co-factors controlled the expression of uPA and uPAR, and the inhibition of NF-κB and activator protein-1 (AP-1) suppressed the secretion of uPA, resulting in the inhibition of motility of highly invasive BC cells [39, 100]. Therefore the role of NF-κB pathway downstream of both HER2 and uPAR assumes significance not only due to the part it plays in tumor initiation, metastasis and recurrence of disease condition with increased aggressiveness, but also from reports that implicate NF-κB to the expansion of CSCs. The latter finding implicating growth and metastasis of the tumor population in CSCs to be driven by HER2 and uPAR mediated by NF-κB may partially explain the failure of existing treatment strategies to completely eradicate solid tumors [107] and drug resistance. For example, one of the theories suggest that the efficacy of currently available drugs that can only shrink metastatic tumors are usually transient and does not lead to extended patient survival [108-110]. This has been blamed on the acquisition of drug resistance by the cancer cells and the failure to kill CSCs effectively by existing therapies. Therefore, the activation of non-canonical pathways through PKC and Src and canonical pathway mediated by NF-κB not only has implications in maintaining constitutive HER2 and uPAR overexpression and hence tumor invasiveness, but also could play a significant role in development of drug resistance.
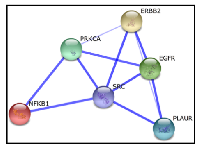
As discussed before, both HER2 and uPAR appear to have functional interactions with downstream intracellular common oncogenic players such as Src, PKCα, and NF-κB. Hence, to further confirm these potential functional associations we used the interaction network database STITCH 4.0 [111] with multiple proteins option using input genes PLAUR, ERBB2, PRKCA, NFKB1, and SRC. All six input genes formed a single protein functional interaction network (Figure 2). This data analysis converges with previous findings implicating these molecules as critical factors in HER2 and uPAR-mediated invasion and metastasis of BC.
EGFR – A preferred dimerization partner of HER2 and an essential signal transducer for uPAR
Another important observation from the STITCH protein interaction network (Figure 2) is the strong association of EGFR with both HER2 and uPAR. Whilst several studies have shown HER2 to be the preferred dimerization partner of other HER family members [112], co-expression of HER2 with EGFR has been shown to induce a synergistic transforming effect on rodent fibroblasts [113]. A recent study has also found EGFR overexpression to be a poor prognostic factor in HER2-positive primary BC [114]. On the other hand, EGFR has also been demonstrated to mediate uPAR/integrin/fibronectin (FN) induced growth pathway leading to the in vivo proliferation of HEp3 human carcinoma [115]. This was further confirmed in a study by Jo et al. [116], where they found EGFR to be an essential component for the transduction of signals from uPAR to ERK in cells that express EGFR. More studies investigating the interactions between EGFR and uPAR followed. For example, Guerrero et al. [88] showed that in mammary epithelial MCF-7 cells expressing low levels of uPAR, stimulation of uPAR with the amino-terminal fragment (ATF) of urokinase devoid of proteolytic activity transactivated the EGFR through a mechanism involving Src and a metalloproteinase leading to cellular invasion. Monaghan-Benson et al. [117] found that binding of P25, a uPAR ligand, to uPAR causes an Src-dependent transactivation of EGFR and promotes the formation of EGFR-β1 complexes leading to upregulation of fibronectin matrix assembly. Jo et al. [118] further reported that uPAR is required for EGF-induced cell growth in MDA-MB 231 breast cancer cells and murine embryonic fibroblasts (MEFs) through Tyr-845 phosphorylation of EGFR and activation of STAT5b. D´Alessio et al. [119] found that mouse keratinocytes deficient for uPAR failed to produce and secrete EGFR-dependent laminin-5, affecting adhesion and migration properties in vitro and wound healing in vivo. Hu et al. [120] demonstrated uPAR to be a highly significant crosstalk molecule that is necessary for the activation of signal transducer and activator of transcription 5b (STAT5b), a recently identified downstream effector of EGFRvIII [121], in glioblastoma multiforme cells. A very recent study by Kozlova et al., [122] reported a uPA-uPAR mediated attenuation of the mitogenic effect of EGF on cellular proliferation, invasion and motility in MCF-7 and MDA-MB-231 breast cancer cells. Though interesting, more studies are needed to confirm in clinical specimens the role of uPA as a negative modulator of EGF-dependent cellular proliferation and motility.Common regulatory transcriptional factors of HER2 and PLAUR
To identify the common transcription factors that have propensity to regulate both HER2 and PLAUR gene expression in HER2-positive breast carcinoma, we submitted their gene symbols into GEMS launcher software. The analyzer identified V$ETSF (Ets family of transcription factors) and V$KLFS (Kruppel-like family of transcription factors) as the common transcription factor families (Figure 3) that bind to and regulate HER2 and PLAUR.
ETS members are transcriptional targets of HER2 and uPAR signaling
The ETS family of transcription factors are defined by a conserved DNA binding domain. This domain forms a winged helix-turn-helix structural motif [123]. Many ETS factors are shown to be dysregulated in BC such as ETS1 (v-ets avian erythroblastosis virus E26 oncogene homolog 1), ETS2 (v-ets avian erythroblastosis virus E26 oncogene homolog 2) and PEA3 (Polyomavirus enhancer activator 3) [124]. A handful of studies have addressed the ETS transcription factors mediated regulation of HER2 and uPAR signaling. ETS proteins have been implicated as downstream factors of HER2 signaling [125] and, at a clinical level, ETS proteins have been shown to associate with breast tumor disease progression and metastasis [74, 126]. These MAP kinase-dependent transcription factors interact with a multitude of co-regulatory partners to elicit a biological process [125, 127]. For example, studies by Myers et al., [75] found that Src-1 is a functional coactivator of ETS-2. Al-azawi et al., [74] reported strong associations between the transcription factor, ETS-2 and its coactivator Src-1 (P<0.01) and the target gene myc (P<0.0001) in a cohort of BC patients with locally advanced disease. On the other hand, PKCα was also found to regulate ETS1 activity as a downstream transcriptional factor in invasive BC cells [128]. On examining the PKC expression in a variety of BC cell lines, Lindermann et al., [128] found that the protein level of PKCα was much higher in ETS1-expressing MDA-MB-231 and MDA-MB-435 BC cells than in ETS1-deficient MCF-7 and SK-BR3 cells, whereas PKCα-deficient MCF-7 cells do not support ETS1-induced activation of the PTHrP P3 promoter strongly suggesting that PKCα may be important for ETS1 activity. To follow it up, studies also found that attenuation of endogenous PKCα expression (siPalpha) by RNA interference leads to reduced ETS1 protein expression in a variety of cancer cells suggesting that ETS1 serves as an effector for PKCα to fulfil certain functions in cancer cells [129].
KLF transcription factors downstream of Src, PKCα, and NF-κB
KLFs are a diverse family of Zinc finger containing DNA binding transcription factors. Currently, 17 KLFs are known in mammals. They have a carboxy terminal DNA binding domain with three Zn fingers. Zn fingers bind to GC rich DNA sequences [130, 131]. Several KLFs are altered or elevated in cancer [131]. In BC, KLF2, KLF4, KLF5, KLF6, KLF8, KLF10 and KLF17 have been found to be altered [130, 132, 133]. For a detailed understanding KLF family members and its context dependent functions, refer to Tetreault et al., [133]. KLF4 expression is associated with BC progression and KLF4 mRNA and protein are overexpressed in up to 70% of BCs [134, 135]. Increased nuclear expression of KLF4 is considered to be associated with the aggressiveness of BC phenotypes [135]. However, the precise role of KLF4 in transcriptional regulation of both the PLAUR and HER2 in BC is yet to be examined in detail. Another KLF member, KLF8 promotes human BC cell invasion and metastasis by transcriptionally repressing cadherin 1 (CDH1) and transactivating matrix metallopeptidase (MMP9) [136, 137], and high expression of KLF8 predicts a poor prognosis in human cancers [133]. KLF10, on the other hand, transcriptionally represses EGFR and inhibits invasion and metastasis in vitro and in an orthotopic mouse tumor model [138], and KLF10 loss is downregulated in invasive human BC [133, 139]. Also, initial studies revealed KLF5 to be a potential tumor suppressor gene in BC [140], however, a recent study found that patients with a higher KLF5 expression have shorter disease-free and OS than patients with a lower KLF5 expression [132, 141]. It was recently shown that reduction or absence of KLF6 abrogates the negative control of BC cell proliferation triggered by ER–alpha through the signaling pathway mediated by c-Src and Akt activation [142]. In other words, cytoplasmic KLF6 is able to interact with c-Src protein and thereby interferes with ER–alpha-mediated cell growth of BC cells.
HER2 and uPAR – Correlative markers and potential dual drug targets
It has been proposed that amplification of a single chromosomal region (for example, HER2) may destabilize the tumor genome, thereby facilitating the amplification of an additional loci [68, 143] (for example, PLAUR). If these amplification combinations were to exist, breast tumors harbouring HER2 gene amplification can be assumed to acquire subsequent amplification of the PLAUR gene at a later stage of tumor development, thereby allowing the tumor cells to acquire the ability to invade the surrounding tissues and spread to distant sites of the body [68]. Regulation of PLAUR and HER2 in advanced BCs by common oncogenic players as evident from the preclinical evidence, STITCH database analysis (Figure 2), and common transcriptional factor binding sites (Figure 3) confirms the role of Src, PKCα, and NF-κB signaling downstream of HER2 and uPAR in altering the amplification status of PLAUR and HER2. Therefore, in HER2-positive early-stage aggressive breast carcinoma, it could be likely that the hyper-activation of common oncogenic players like Src, PKCα and NF-κB may act in a non-canonical mechanism either independently or in a concerted manner to upregulate the amplification status of PLAUR and/or HER2 through activation of common transcription factors such as ETS and KLF (Figure 4A & B). This partly explains the existence of PLAUR amplification in CTCs from patients with HER2-positive BC [33] and is consistent with previous reports that showed marked preference for amplification of both HER2 and PLAUR genes to occur in the same CTC in HER2-amplified tumors [144, 145]. Since CTCs have been reported recently as precursors and contribute significantly to BC metastasis [146], high HER2 and PLAUR co-amplification can be expected to be seen only during the later stages of malignant tumor development (CTCs to distant metastases stage). Therefore, upregulation of HER2 and PLAUR by common oncogenic players can be attributed as being specific to an early-stage aggressive breast carcinoma subtype.
According to the Tumor Marker Utility Grading System, uPA and PAI-1 invasion markers along with HER2 are still the most dominant independent novel prognostic factors that have reached the highest level of evidence for clinical utility in BC [4, 147, 148]. It is evident from previous studies [149, 150] and Figure 1B, that uPA is highly correlated with uPAR in BC. Various studies have shown uPA/PAI-1 and HER2 as independent prognostic and predictive markers for DFS and on aggressive outcome in lymph node-negative BC [6, 151-154]. At the same time, uPA and PAI-1 mRNA expression have been shown to have a strong association with shorter DFS (p = 0.013 for PAI-1, p = 0.001 for uPA) in HER2-positive BC patients [35, 155]. Currently, the main clinical relevance of uPA/PAI-1 as prognostic biomarkers is in the identification of lymph node-negative patients with HER-2-negative tumors for adjuvant chemotherapy [152]. However, the combined clinical relevance of HER2 and uPA/PAI-1 can significantly contribute towards optimal decision making in the selection of patients with primary BC for various treatment strategies. Since uPA and uPAR strongly correlate (Figure 1B), and patients with tumors expressing high uPA, high uPAR, and/or high PAI-1 levels show a significantly shorter RFS and OS compared to patients with low levels of their expression [150], the assessment of both markers together with HER2 and PAI-1 in BCs will enable clinicians to accurately predict the disease outcome and to identify in early stage patients, who will benefit from combined therapies.
In the light of the HER2 and uPAR cooperativity and the common regulatory signaling pathway downstream of HER2 and uPAR in advanced breast carcinoma (Figure 4B), the correlative co-expression pattern of HER2 and uPAR definitely has the potential to act as synergistic targets for therapeutic intervention. This suggestion is well supported by studies done by Li et al., [157] who found that downregulation of HER2/uPAR individually at the cell surface, leads to decreased ERK activity and this effect maximizes upon downregulation of both receptors simultaneously indicating a synergistic effect on BC cells. Li et al., [157] further demonstrated that RNA interference (RNAi) depletion of either HER2 or uPAR suppressed cell growth and induced cell apoptosis, and these effects were significantly enhanced in cells depleted of both HER2 and uPAR. Moreover, downregulation of uPAR using RNAi synergized with trastuzumab to suppress the growth and induce apoptosis of SKBR3 and ZR751 cells and this effect was also evident in the mechanistic analysis where uPAR RNAi significantly enhanced the effect of trastuzumab on inhibition of MAPK signal pathways. This recent finding makes these receptors potential targets for combinatorial therapies using either trastuzumab and uPAR antagonists or selective small molecules or antibody-drug conjugates to achieve inhibition of HER2 and uPAR. It can be expected that simultaneous targeting of HER2 and uPAR, the cooperativity of which this review discusses and substantiates to contribute to the metastatic phenotype of HER2-positive BC, may possibly convert a cell´s phenotype from tumorigenic to dormant or prolong their dormant state with less adverse side effects.
CONCLUSION
Previously, the model of metastasis was explained in terms of rare subpopulations of cells within the primary tumor that acquire advantageous genetic alterations over a period of time, enabling these cells to metastasize and form new solid tumors at distant sites [158]. This genetic selection model of metastasis was debated for some time by various groups [159-161], before the emergence of gene expression profiling data [162-164]. Studies based on DNA microarrays reported that primary breast tumors can be distinguished by their gene expression profile for their metastatic potential. This implies that genetic mutations determine metastatic behavior at early stages of tumorigenesis [165]. This review confirms previous knowledge and substantiates non-canonical mechanisms contributing to the cooperativity between HER2 and uPAR in advanced BC. This process involves various other downstream molecules including Src/PKCα/NF-κB leading to the activation of transcriptional factors such as ETS or KLF that contribute to the aggressiveness of HER2-positive breast carcinoma phenotype and possibly cause feed-back resistance mechanism to HER2 targeted therapy. Based on the critical nature of cooperativity between HER2 and uPAR in advanced HER2-positive breast carcinoma, this review also stresses the importance of targeting simultaneously HER2 and uPAR to improve personalized treatment modalities of newly diagnosed patients.
Abbreviations
BC, Breast cancer; FDA, Food and Drug Administration; uPAR, urokinase plasminogen activator receptor; uPA, urokinase plasminogen activator; PAI-1, plasminogen activator inhibitor type 1; RFS, Relapse-free survival; HER2, Human epidermal growth factor receptor type 2; DFS, disease-free survival; CTCs, circulating tumor cells; DTCs, disseminated tumor cells; ER, estrogen receptor; PR, progesterone receptor; NF-κB, nuclear factor-kappaB; CSCs, cancer stem cells; GPI, glycosyl phosphatidylinositol; GPCR, G-protein-coupled receptor; Lck, lymphocyte protein tyrosine kinase; Hck, haematopoietic cell kinase; FAK, focal adhesion kinase; ERK, extracellular-signal-regulated kinase; MAPK, mitogen-activated protein kinase; MFS, metastases-free survival; HDPP, HER2-derived prognostic predictor; OS, overall survival; DMFS, distant metastasis-free survival; NKI, Nederlands Kanker Instituut; FFPE, formalin-fixed and paraffin-embedded; RPPA, reverse phase protein microarray; PKCα, Protein kinase Cα; Src, steroid receptor co-activator; NCOA1/NCOA2/NCOA2, nuclear receptor co-activator 1/2/3; TIF2, transcriptional intermediary factor-2; GRIP1, glucocorticoid receptor interacting protein-1; AIB1, amplified in BC-1; ACTR, activator of retinoid and thyroid receptors; PLCγ, phospholipase-γ; siRNA, small interfering RNA; ALDH, aldehyde dehydrogenase; AP-1, activator protein-1; EGFR, epidermal growth factor receptor; FN, fibronectin; ATF, amino-terminal fragment; STAT5b, signal transducer and activator of transcription 5b; ETSF, Ets family of transcription factors; KLFS, Kruppel-like family of transcription factors; PEA3, Polyomavirus enhancer activator 3; CDH1, cadherin 1; MMP9, Matrix metallopeptidase; TCGA, The Cancer Genome Atlas; RNAi, RNA interference.
ACKNOWLEDGMENTS
We acknowledge a University of Wollongong Vice-Chancellor Postdoctoral Fellowship and a Cure Cancer Australia Foundation Project Grant (APP1045831) to Dr. Kara L. Vine. A Cancer Institute NSW Fellowship to Prof. Marie Ranson is also gratefully acknowledged. We also thank Mr. Sven Giese (Ph.D candidate, University of Edinburgh) for assistance with Figure 1A&B (HER2 and PLAUR correlation curves). A particular acknowledgment goes to Dr. Vincent Vuaroqueaux who supervised the detection of PLAUR and HER2 mRNA transcriptional values at the former Stiftung Tumorbank Basel. We also thank Prof. Mattias Belting for critical reading of the manuscript.
CONFLICT OF INTEREST
No conflict of interests was disclosed by the authors.

- 1. Targeted therapies for breast cancer. The Journal of Clinical Investigation. 2011; 121: 3797-3803. https://doi.org/10.1172/JCI57152. [PubMed].
- 2. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012; 109: 2724-9. https://doi.org/10.1073/pnas.1018854108. [PubMed].
- 3. The role of chemotherapy and targeted agents in patients with metastatic breast cancer. European Journal of Cancer. 2011; 47: S38-S47. https://doi.org/10.1016/S0959-8049(11)70145-9. [PubMed].
- 4. Breast cancer metastasis: markers and models. Nature Reviews Cancer. 2005; 5: 591-602. https://doi.org/10.1038/nrc1670. [PubMed].
- 5. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. Journal of Clinical Oncology. 2007; 25: 5287-5312. https://doi.org/10.1200/JCO.2007.14.2364. [PubMed].
- 6. Invasion factors uPA/PAI-1 and HER2 status provide independent and complementary information on patient outcome in node-negative breast cancer. Journal of Clinical Oncology. 2003; 21: 1022-1028. https://doi.org/10.1200/JCO.2003.04.170. [PubMed].
- 7. Lund IK, Illemann M, Thurison T, Christensen IJ, and Høyer-Hansen G. uPAR as anti-cancer target: evaluation of biomarker potential, histological localization, and antibody-based therapy. Current Drug Targets. 2011; 12: 1744-1760. https://doi.org/10.2174/138945011797635902. [PubMed].
- 8. Human breast cancer: correlation of relapse and survival with amplification of the HER2/neu oncogene. Science. 1987; 235: 177-182. https://doi.org/10.1126/science.3798106. [PubMed].
- 9. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989; 244: 707-712. https://doi.org/10.1126/science.2470152. [PubMed].
- 10. Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, Hung G, Robinson RA, Harris C, El-Naggar A, Slamon DJ, Phillips RN, Ross JS, et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. Journal of Clinical Oncology. 1997; 15: 2894-2904. https://doi.org/10.1200/JCO.1997.15.8.2894. [PubMed].
- 11. Evaluation of HER-2/neu gene amplification and over-expression: comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. Journal of Clinical Oncology. 2002; 20: 3095–3105.
- 12. Anti-human epidermal growth factor receptor 2 monoclonal antibody therapy for breast cancer. British Journal of Surgery. 2002; 89: 262-271. https://doi.org/10.1046/j.0007-1323.2001.02022.x. [PubMed].
- 13. Amplification of erbB2 and erbB2 expression are superior to estrogen receptor status as risk factors for distant recurrence in pT1N0M0 breast cancer: a nationwide population based study. Clinical Cancer Research. 2003; 9: 923–930. [PubMed].
- 14. Esteva FJ, Cheli CD, Fritsche H, Fornier M, Slamon D, Thiel RP, Luftner D, and Ghani F. Clinical utility of serum HER2/neu in monitoring and prediction of progression-free survival in metastatic breast cancer patients treated with trastuzumab-based therapies. Breast Cancer Research. 2005; 7: R436–R443. https://doi.org/10.1186/bcr1020. [PubMed].
- 15. Prognostic and predictive significance of ErbB-2 breast tumor levels measured by enzyme immunoassay. Journal of Clinical Oncology. 2001; 19: 645-656. https://doi.org/10.1200/JCO.2001.19.3.645. [PubMed].
- 16. Di Fiore PP, Pierce JH, Kraus MH, Segatto O, King CR, and Aaronson SA. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 1987; 237: 178-182. https://doi.org/10.1126/science.2885917. [PubMed].
- 17. Liu Y, el-Ashry D, Chen D, Ding IY, and Kern FG. MCF-7 breast cancer cells overexpressing transfected c-erbB-2 have an in vitro growth advantage in estrogen-depleted conditions and reduced estrogen-dependence and tamoxifen-sensitivity in vivo. Breast Cancer Research and Treatment. 1995; 34: 97-117. https://doi.org/10.1007/BF00665783. [PubMed].
- 18. Overexpression of the cerbB- 2 gene enhanced intrinsic metastasis potential in human breast cancer cells without increasing their transformation abilities. Cancer Research. 1997; 57: 1199–1205.
- 19. Differential modulation of proliferation, matrix metalloproteinase expression and invasion of human head and neck squamous carcinoma cells by c-erbB ligands. Clinical and Experimental Metastasis. 1999; 17: 631-639. https://doi.org/10.1023/a:1006751016860. [PubMed].
- 20. ‘Accidental’ anti-angiogenic drugs. anti-oncogene directed signal transduction inhibitors and conventional chemotherapeutic agents as examples. European Journal of Cancer. 2000; 36: 1248-1257.
- 21. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nature Cell Biology. 2001; 3: 785-792. https://doi.org/10.1038/ncb0901-785. [PubMed].
- 22. HER2/Neu signaling and therapeutic approaches in breast cancer. Breast Disease. 2002; 15: 13-24. https://doi.org/10.3233/bd-2002-15103. [PubMed].
- 23. Cancer cell motility--on the road from c-erbB-2 receptor steered signaling to actin reorganization. Experimental Cell Research. 2002; 272: 93-108. https://doi.org/10.1006/excr.2001.5385. [PubMed].
- 24. Induction of cancer cell migration by epidermal growth factor is initiated by specific phosphorylation of tyrosine 1248 of c-erbB-2 receptor via EGFR. FASEB Journal. 2002; 16: 1823-1825. https://doi.org/10.1096/fj.02-0096fje. [PubMed].
- 25. Role of HER2/neu in tumor progression and therapy. Cellular and Molecular Life Sciences. 2004; 61: 2965-2978. https://doi.org/10.1007/s00018-004-4277-7. [PubMed].
- 26. c-erbB-2 expression and response to adjuvant therapy in women with nodepositive early breast cancer. New England Journal of Medicine. 1994; 330: 1260–1266. https://doi.org/10.1056/NEJM199405053301802. [PubMed].
- 27. erbB-2, p53, and efficacy of adjuvant therapy in lymph node-positive breast cancer. Journal of the National Cancer Institute. 1998; 90: 1346–1360. https://doi.org/10.1093/jnci/90.18.1346. [PubMed].
- 28. erbB-2 and response to doxorubicin in patients with axillary lymph nodepositive, hormone receptor-negative breast cancer. Journal of the National Cancer Institute. 1998; 90: 1361-1370.
- 29. Stability of HER-2/neu expression over time and at multiple metastatic sites. Journal of the National Cancer Institute. 1993; 85: 1230–1235. https://doi.org/10.1093/jnci/85.15.1230. [PubMed].
- 30. Nahta R, Yu D, Hung M, Hortobagyi GN, and Esteva FJ. Mechanisms of Disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nature Clinical Practice Oncology. 2006a; 3: 269-280. https://doi.org/10.1038/ncponc0509. [PubMed].
- 31. HER2 therapy: Molecular mechanisms of trastuzumab resistance. Breast Cancer Research. 2006b; 8: 215. https://doi.org/10.1186/bcr1612. [PubMed].
- 32. Pierga J-Y, Bonneton C, Magdelénat H, Vincent-Salomon A, Nos C, Boudou E, Pouillart P, Thiery J-P, and Cremoux Pd. Real-time quantitative PCR determination of urokinase-type plasminogen activator receptor (uPAR) expression of isolated micrometastatic cells from bone marrow of breast cancer patients. International Journal of Cancer. 2005; 114: 291-298. https://doi.org/10.1002/ijc.20698. [PubMed].
- 33. UPAR and HER2 gene status in individual breast cancer cells from blood and tissues. Proceedings of the National Academy of Sciences of the United States of America. 2006; 103: 17361-17365. https://doi.org/10.1073/pnas.0608113103. [PubMed].
- 34. Upregulation and activation of PKC by ErbB2 through Src promotes breast cancer cell invasion that can be blocked by combined treatment with PKC and Src inhibitors. Oncogene. 2006; 25: 3286–3295. https://doi.org/10.1038/sj.onc.1209361. [PubMed].
- 35. Urban P, Vuaroqueaux V, Labuhn M, Delorenzi M, Wirapati P, Wight E, Senn HJ, Benz C, Eppenberger U, and Eppenberger-Castori S. Increased expression of urokinase-type plasminogen activator mRNA determines adverse prognosis in ErbB2-positive primary breast cancer. Journal of Clinical Oncology. 2006; 24: 4245-4253. https://doi.org/10.1200/JCO.2005.05.1912. [PubMed].
- 36. Identification of Subtypes in Human Epidermal Growth Factor Receptor 2–Positive Breast Cancer Reveals a Gene Signature Prognostic of Outcome. Journal of Clinical Oncology. 2010; 28: 1813-1820. https://doi.org/10.1200/JCO.2009.22.8775. [PubMed].
- 37. Profiling signalling pathways in formalin-fixed and paraffin-embedded breast cancer tissues reveals cross-talk between EGFR, HER2, HER3 and uPAR. Journal of Cellular Physiology. 2012; 227: 204-212. https://doi.org/10.1002/jcp.22718. [PubMed].
- 38. Mesenchymal Phenotype of CTC-Enriched Blood Fraction and Lymph Node Metastasis Formation Potential. PLoS ONE. 2014; 9: e93901. https://doi.org/10.1371/journal.pone.0093901. [PubMed].
- 39. The role of breast cancer stem cells in metastasis and therapeutic implications. The American Journal of Pathology. 2011b; 179: 2-11. https://doi.org/10.1016/j.ajpath.2011.03.005. [PubMed].
- 40. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, et al. Molecular portraits of human breast tumors. Nature. 2000; 406: 747-752. https://doi.org/10.1038/35021093. [PubMed].
- 41. Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America 2001; 98: 10869-10874. https://doi.org/10.1073/pnas.191367098. [PubMed].
- 42. Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences of the United States of America 2003; 100: 8418-8423. https://doi.org/10.1073/pnas.0932692100. [PubMed].
- 43. Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 2006; 7. https://doi.org/10.1186/1471-2164-7-96. [PubMed].
- 44. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of Clinical Oncology. 2009; 27: 1160-1167. https://doi.org/10.1200/JCO.2008.18.1370. [PubMed].
- 45. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. The Journal of Pathology. 2010; 220: 263-280. https://doi.org/10.1002/path.2648. [PubMed].
- 46. Claudin-low breast cancers: clinical, pathological, molecular and prognostic characterization. Molecular Cancer. 2014; 13: 228. https://doi.org/10.1186/1476-4598-13-228. [PubMed].
- 47. Deconstructing the molecular portraits of breast cancer. Molecular Oncology. 2011; 5: 5-23. https://doi.org/10.1016/j.molonc.2010.11.003. [PubMed].
- 48. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Research. 2010; 12: R68. https://doi.org/10.1186/bcr2635. [PubMed].
- 49. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treatment Reviews. 2012; 38: 698-707. https://doi.org/10.1016/j.ctrv.2011.11.005. [PubMed].
- 50. Metastatic Behavior of Breast Cancer Subtypes. Journal of Clinical Oncology. 2010; 28: 3271-3277. https://doi.org/10.1200/JCO.2009.25.9820. [PubMed].
- 51. Biological subtypes of breast cancer: current concepts and implications for recurrence patterns. Quarterly Journal of Nuclear Medicine and Molecular Imaging. 2013; 57: 312-321. [PubMed].
- 52. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. New England Journal of Medicine. 2001; 344: 783-792. https://doi.org/10.1056/NEJM200103153441101. [PubMed].
- 53. Metastatic patterns of breast cancer subtypes: What radiologists should know in the era of personalized cancer medicine. Clinical Radiology. 2015; 70: 1-10. https://doi.org/10.1016/j.crad.2014.08.015. [PubMed].
- 54. Personalizing extent of breast cancer surgery according to molecular subtypes. The Breast. 2013; 22, Supplement 2: S106-S109. https://doi.org/10.1016/j.breast.2013.07.020. [PubMed].
- 55. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nature Reviews Cancer. 2007; 7: 389–397. https://doi.org/10.1038/nrc2127. [PubMed].
- 56. ErbB receptors and cell polarity: new pathways and paradigms for understanding cell migration and invasion. Experimental Cell Research. 2009; 315: 707–716. https://doi.org/10.1016/j.yexcr.2008.10.034. [PubMed].
- 57. ErbB receptors and signaling pathways in cancer. Current Opinion in Cell Biology. 2009; 21: 177-184. https://doi.org/10.1016/j.ceb.2008.12.010. [PubMed].
- 58. Ranson M and Andronicos NM. Plasminogen binding and cancer: promises and pitfalls. Frontiers in Bioscience 2003; 8: s294-304. https://doi.org/10.2741/1044. [PubMed].
- 59. Regulation of cell migration and invasion by specific modules of uPA: mechanistic insights and specific inhibitors. Current Drug Targets. 2011; 12: 1761-1771. https://doi.org/10.2174/138945011797635777. [PubMed].
- 60. The urokinase-type plasminogen activator and the generation of inhibitors of urokinase activity and signaling. Current Pharmaceutical Design. 2011; 17: 1944 - 1961. https://doi.org/10.2174/138161211796718143. [PubMed].
- 61. The Urokinase Receptor Is Expressed in Invasive Breast Cancer but not in Normal Breast Tissue. Cancer Research. 1994; 54: 861-866. [PubMed].
- 62. Targeting uPAR with Antagonistic Recombinant Human Antibodies in Aggressive Breast Cancer. Cancer Research. 2013; 73: 2070-2081. https://doi.org/10.1158/0008-5472.CAN-12-3526. [PubMed].
- 63. The prognostic value of the topographic distribution of uPAR expression in invasive breast carcinomas. Cancer Letters. 2007; 246: 262-267. https://doi.org/10.1016/j.canlet.2006.03.003. [PubMed].
- 64. The Many Spaces of uPAR: Delivery of Theranostic Agents and Nanobins to Multiple Tumor Compartments through a Single Target. Theranostics. 2013; 3: 496-506. https://doi.org/10.7150/thno.4953. [PubMed].
- 65. Cancer invasion and tissue remodeling-cooperation of protease systems and cell types. acta pathologica, microbiologica, et immunologica Scandinavica. 1999; 107: 120-127. https://doi.org/10.1111/j.1699-0463.1999.tb01534.x. [PubMed].
- 66. Urokinase type plasminogen activator receptor expression in colorectal neoplasms. Gut. 1998; 43: 798-805. https://doi.org/10.1136/gut.43.6.798. [PubMed].
- 67. Jo M, Eastman BM, Webb DL, Stoletov K, Klemke R, and Gonias SL. Cell Signaling by Urokinase-type Plasminogen Activator Receptor Induces Stem Cell–like Properties in Breast Cancer Cells. Cancer Research. 2010; 70: 8948-8958. https://doi.org/10.1158/0008-5472.CAN-10-1936. [PubMed].
- 68. Bempt IV, Drijkoningen M, and De Wolf-Peeters C. The complexity of genotypic alterations underlying HER2-positive breast cancer: an explanation for its clinical heterogeneity. Current Opinion in Oncology. 2007; 19: 552-557. https://doi.org/10.1097/CCO.0b013e3282f0ad8e. [PubMed].
- 69. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery. 2012; 2: 401-404. https://doi.org/10.1158/2159-8290.CD-12-0095. [PubMed].
- 70. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science Signaling. 2013; 6: pl1. https://doi.org/10.1126/scisignal.2004088. [PubMed].
- 71. Regulation of motility and protease expression in PKC-mediated induction of MCF-7 breast cancer cell invasiveness. Experimental Cell Research. 1999; 247: 105–113. https://doi.org/10.1006/excr.1998.4336. [PubMed].
- 72. Combination of an SRC kinase inhibitor with a novel pharmacological antagonist of the urokinase receptor diminishes in vitro colon cancer invasiveness. Clinical Cancer Research. 2004; 10: 1545–1555. https://doi.org/10.1158/1078-0432.ccr-1565-02. [PubMed].
- 73. Review of the in vivo functions of the p160 steroid receptor coactivator family. Molecular Endocrinology. 2003; 17: 1681–1692. https://doi.org/10.1210/me.2003-0116. [PubMed].
- 74. Al-azawi D, Ilroy MM, Kelly G, Redmond AM, Bane FT, Cocchiglia S, Hill AD, and Young LS. Ets-2 and p160 proteins collaborate to regulate c-Myc in endocrine resistant breast cancer. Oncogene 2008; 27: 3021-3031. https://doi.org/10.1038/sj.onc.1210964. [PubMed].
- 75. Myers E, Hill AD, Kelly G, McDermott EW, O’Higgins NJ, Buggy Y, and Young LS. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, andNCoRin breast cancer. Clinical Cancer Research 2005; 11: 2111-2122. https://doi.org/10.1158/1078-0432.CCR-04-1192. [PubMed].
- 76. Concerted activation of ETS protein ER81 by p160 coactivators,the acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. Journal of Biological Chemistry. 2004; 279: 14909–14916. https://doi.org/10.1074/jbc.M400036200. [PubMed].
- 77. Fleming FJ, Myers E, Kelly G, Crotty TB, McDermott EW, O’Higgins NJ, Hill AD, and Young LS. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. Journal of Clinical Pathology 2004; 57: 1069-1074. https://doi.org/10.1136/jcp.2004.016733. [PubMed].
- 78. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Molecular and Cellular Biology. 2008; 28: 5937–5950. https://doi.org/10.1128/MCB.00579-08. [PubMed].
- 79. Louie MC, Revenko AS, Zou JX, Yao J, and Chen HW. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Molecular and Cellular Biology. 2006; 26: 3810-3823. https://doi.org/10.1128/MCB.26.10.3810-3823.2006. [PubMed].
- 80. Stimulation of steroid receptor coactivator-3 (SRC-3) gene overexpression by a positive regulatory loop of E2F1 and SRC-3. Molecular Endocrinology. 2006; 20: 3105-3119. https://doi.org/10.1210/me.2005-0522. [PubMed].
- 81. Genetic screening reveals an essential role of p27kip1 in restriction of breast cancer progression. Cancer Research. 2007; 67: 8032–8042. https://doi.org/10.1158/0008-5472.CAN-07-0083. [PubMed].
- 82. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proceedings of the National Academy of Sciences of the United States of America. 2009; 106: 151-156. https://doi.org/10.1073/pnas.0808703105. [PubMed].
- 83. Fleming FJ, Hill AD, McDermott EW, O’Higgins NJ, and Young LS. Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. Journal of Clinical Endocrinology and Metabolism. 2004; 89: 375-383. https://doi.org/10.1210/jc.2003-031048. [PubMed].
- 84. Activation of Src family kinases in Neu-induced mammary tumors correlates with their association with distinct sets of tyrosine phosphorylated proteins in vivo. Oncogene. 1995; 11: 1801–1810. [PubMed].
- 85. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004; 6: 117-127. https://doi.org/10.1016/j.ccr.2004.06.022. [PubMed].
- 86. ErbB2 promotes Src synthesis and stability: novel mechanisms of Src activation that confer breast cancer metastasis. Cancer Research. 2005; 65: 1858–1867. https://doi.org/10.1158/0008-5472.CAN-04-2353. [PubMed].
- 87. Xu Y, Hu B, Qin L, Zhao L, Wang Q, Wang Q, Xu Y, and Jiang J. SRC-1 and Twist1 Expression Positively Correlates with a Poor Prognosis in Human Breast Cancer. International Journal of Biological Sciences. 2014; 10: 396-403. https://doi.org/10.7150/ijbs.8193. [PubMed].
- 88. EGF receptor transactivation by urokinase receptor stimulus through a mechanism involving Src and matrix metalloproteinases. Experimental Cell Research. 2004; 292: 201-208. https://doi.org/10.1016/j.yexcr.2003.08.011. [PubMed].
- 89. Protein kinase C isozymes and the regulation of diverse cell responses. American Journal of Physiology - Lung Cellular and Molecular Physiology. 2000; 279: L429–L438. https://doi.org/10.1152/ajplung.2000.279.3.L429. [PubMed].
- 90. PKC at a glance. Journal of Cell Science. 2004; 117: 131-132. https://doi.org/10.1242/jcs.00982. [PubMed].
- 91. Protein kinase C and its isoforms in human breast cancer cells: relationship to the invasive phenotype. International Journal of Oncology. 1998; 12: 1349–1354. https://doi.org/10.3892/ijo.12.6.1349. [PubMed].
- 92. Masur K, Lang K, Niggemann B, Zanker KS, and Entschladen F. High PKC alpha and low E-cadherin expression contribute to high migratory activity of colon carcinoma cells. Molecular Biology of the Cell 2001; 12: 1973-1982. https://doi.org/10.1091/mbc.12.7.1973. [PubMed].
- 93. Protein kinase C alpha expression in breast and ovarian cancer. Oncology. 2004; 67: 1-10. https://doi.org/10.1159/000080279. [PubMed].
- 94. The link between PKCalpha regulation and cellular transformation. Immunology Letters. 2005; 96: 155–162. https://doi.org/10.1016/j.imlet.2004.08.013. [PubMed].
- 95. MCF-7 breast cancer cells transfected with protein kinase C-alpha exhibit altered expression of other protein kinase C isoforms and display a more aggressive neoplastic phenotype. The Journal of Clinical Investigation. 1995; 95: 1906–1915. https://doi.org/10.1172/JCI117872. [PubMed].
- 96. PKCα expression is a marker for breast cancer aggressiveness. Molecular Cancer. 2010; 9. https://doi.org/10.1186/1476-4598-9-76. [PubMed].
- 97. Oncogenic forms of the neu/HER2 tyrosine kinase are permanently coupled to phospholipase C gamma. EMBO J. 1991; 10: 2077–2086. [PubMed].
- 98. Protein Kinase Cα Determines HER2 Fate in Breast Carcinoma Cells with HER2 Protein Overexpression without Gene Amplification. Cancer Research. 2007; 67: 5308-5317. https://doi.org/10.1158/0008-5472.CAN-06-3936. [PubMed].
- 99. Induction of cell migration by pro-urokinase binding to its receptor: possible mechanism for signal transduction in human epithelial cells. Journal of Cell Biology. 1994; 126: 259-270. https://doi.org/10.1083/jcb.126.1.259. [PubMed].
- 100. Sliva D, English D, Lyons D, and Lloyd FPJ. Protein kinase C induces motility of breast cancers by upregulating secretion of urokinase-type plasminogen activator through activation of AP-1 and NF-kappaB. Biochemical and Biophysical Research Communications. 2002; 290: 552-557. https://doi.org/10.1006/bbrc.2001.6225. [PubMed].
- 101. Merkhofer EC, Cogswell P, and Baldwin AS. Her2 activates NF-kappaB and induces invasion through the canonical pathway involving IKKalpha. Oncogene. 2010; 29: 1238-1248. https://doi.org/10.1038/onc.2009.410. [PubMed].
- 102. Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000; 19: 1123–1131. https://doi.org/10.1038/sj.onc.1203412. [PubMed].
- 103. The canonical NF-kappaB pathway governs mammary tumorigenesis in transgenic mice and tumor stem cell expansion. Cancer Research. 2010; 70: 10464–10473. https://doi.org/10.1158/0008-5472.CAN-10-0732. [PubMed].
- 104. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007; 1: 555–567. https://doi.org/10.1016/j.stem.2007.08.014. [PubMed].
- 105. Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, and Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009; 138: 1083-1095. https://doi.org/10.1016/j.cell.2009.06.048. [PubMed].
- 106. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008; 27: 6120–6130. https://doi.org/10.1038/onc.2008.207. [PubMed].
- 107. Treatment of disseminated germ-cell tumors with cisplatin, bleomycin, and either vinblastine or etoposide. New England Journal of Medicine. 1987; 316: 1435-1440. https://doi.org/10.1056/NEJM198706043162302. [PubMed].
- 108. Systematic reviews of chemotherapy and endocrine therapy in metastatic breast cancer. Cancer Treatment Reviews. 2000; 26: 151-168. https://doi.org/10.1053/ctrv.1999.0161. [PubMed].
- 109. High-dose chemotherapy plus autologous bone marrow transplantation for metastatic breast cancer. New England Journal of Medicine. 2000; 342: 1119-1120. https://doi.org/10.1056/NEJM200004133421508. [PubMed].
- 110. Reya T, Morrison SJ, Clarke MF, and Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001; 414: 105-111. https://doi.org/10.1038/35102167. [PubMed].
- 111. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013; 41: D808-D815. https://doi.org/10.1093/nar/gks1094. [PubMed].
- 112. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Molecular and Cellular Biology. 1996; 16: 5276-5287. https://doi.org/10.1128/mcb.16.10.5276. [PubMed].
- 113. Synergistic interaction of p185c-neu and the EGF receptor leads to transformation of rodent fibroblasts. Cell. 1989; 58: 287-292. https://doi.org/10.1016/0092-8674(89)90843-x. [PubMed].
- 114. Prognostic and predictive values of EGFR overexpression and EGFR copy number alteration in HER2-positive breast cancer. British Journal of Cancer. 2014. https://doi.org/10.1038/bjc.2014.556. [PubMed].
- 115. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell. 2002; 1: 445-457. https://doi.org/10.1016/s1535-6108(02)00072-7. [PubMed].
- 116. Epidermal Growth Factor Receptor-dependent and -independent Cell-signaling Pathways Originating from the Urokinase Receptor. Journal of Biological Chemistry. 2003; 278: 1642-1646. https://doi.org/10.1074/jbc.M210877200. [PubMed].
- 117. Monaghan-Benson E and McKeown-Longo PJ. Urokinase-type Plasminogen Activator Receptor Regulates a Novel Pathway of Fibronectin Matrix Assembly Requiring Src-dependent Transactivation of Epidermal Growth Factor Receptor. Journal of Biological Chemistry. 2006; 281: 9450-9459. https://doi.org/10.1074/jbc.M501901200. [PubMed].
- 118. Urokinase receptor primes cells to proliferate in response to epidermal growth factor. Oncogene. 2006; 26: 2585-2594. https://doi.org/10.1038/sj.onc.1210066. [PubMed].
- 119. uPAR-deficient mouse keratinocytes fail to produce EGFR-dependent laminin-5, affecting migration in vivo and in vitro. Journal of Cell Science. 2008; 121: 3922-3932. https://doi.org/10.1242/jcs.037549. [PubMed].
- 120. Crosstalk between the urokinase-type plasminogen activator receptor and EGF receptor variant III supports survival and growth of glioblastoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108: 15984-15989. https://doi.org/10.1073/pnas.1113416108. [PubMed].
- 121. Analysis of Phosphotyrosine Signaling in Glioblastoma Identifies STAT5 as a Novel Downstream Target of ΔEGFR. Journal of Proteome Research. 2011; 10: 1343-1352. https://doi.org/10.1021/pr101075e. [PubMed].
- 122. Urokinase is a negative modulator of Egf-dependent proliferation and motility in the two breast cancer cell lines MCF-7 and MDA-MB-231. Molecular Carcinogenesis. 2015: n/a-n/a. https://doi.org/10.1002/mc.22267. [PubMed].
- 123. Wasylyk B, Hagman J, and Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends in Biochemical Sciences. 1998; 23: 213-216. https://doi.org/10.1016/s0968-0004(98)01211-0. [PubMed].
- 124. Global gene expression analysis identifies PDEF transcriptional networks regulating cell migration during cancer progression. Molecular Biology of the Cell. 2008; 19: 3745–3757. https://doi.org/10.1091/mbc.E08-02-0154. [PubMed].
- 125. Galang CK, García-Ramírez J, Solski PA, Westwick JK, Der CJ, Neznanov NN, Oshima RG, and Hauser CA. Oncogenic Neu/ErbB-2 increases ets, AP-1, and NF-kappaB-dependent gene expression, and inhibiting ets activation blocks Neu-mediated cellular transformation. Journal of Biological Chemistry. 1996; 271: 7992-7998. https://doi.org/10.1074/jbc.271.14.7992. [PubMed].
- 126. Shepherd T and Hassell JA. Role of Ets transcription factors in mammary gland development and oncogenesis. Journal of Mammary Gland Biology and Neoplasia 2001; 6: 129-140. https://doi.org/10.1023/a:1009576801226. [PubMed].
- 127. The ETS-domain transcription factor family. Nature Reviews Molecular Cell Biology. 2001; 2: 827-837. https://doi.org/10.1038/35099076. [PubMed].
- 128. Protein kinase Calpha regulates Ets1 transcriptional activity in invasive breast cancer cells. International Journal of Oncology. 2003; 22: 799-805. [PubMed].
- 129. Ets1 is an effector of protein kinase Calpha in cancer cells. Oncogene. 2005; 24: 650-661. https://doi.org/10.1038/sj.onc.1208234. [PubMed].
- 130. Esophageal squamous cell dysplasia and delayed differentiation with deletion of kruppel-like factor 4 in murine esophagus. Gastroenterology. 2010; 139: 171-181. https://doi.org/10.1053/j.gastro.2010.03.048. [PubMed].
- 131. Krüppel-like factors in cancer progression: three fingers on the steering wheel. Oncotarget. 2014; 5: 29-48. https://doi.org/10.18632/oncotarget.1456. [PubMed].
- 132. Bureau C, Hanoun N, Torrisani J, Vinel JP, Buscail L, and Cordelier P. Expression and Function of Kruppel Like-Factors (KLF) in Carcinogenesis. Current Genomics. 2009; 10: 353-360. https://doi.org/10.2174/138920209788921010. [PubMed].
- 133. Krüppel-like factors in cancer. Nature Reviews Cancer. 2013; 13: 701–713. https://doi.org/10.1038/nrc3582. [PubMed].
- 134. Increase of GKLF Messenger RNA and Protein Expression during Progression of Breast Cancer. Cancer Research. 2000; 60: 6488-6495. [PubMed].
- 135. Nuclear Localization of KLF4 Is Associated with an Aggressive Phenotype in Early-Stage Breast Cancer. Clinical Cancer Research. 2004; 10: 2709-2719. https://doi.org/10.1158/1078-0432.ccr-03-0484. [PubMed].
- 136. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Research. 2007; 67: 7184–7193. https://doi.org/10.1158/0008-5472.CAN-06-4729. [PubMed].
- 137. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011; 30: 1901–1911. https://doi.org/10.1038/onc.2010.563. [PubMed].
- 138. TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Molecular and Cellular Biology. 2012; 32: 50-63. https://doi.org/10.1128/MCB.06152-11. [PubMed].
- 139. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. Journal of Cellular Biochemistry. 1998; 68: 226-236. [PubMed].
- 140. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002; 21: 6567–6572. https://doi.org/10.1038/sj.onc.1205817. [PubMed].
- 141. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clinical Cancer Research. 2006; 12: 2442–2448. https://doi.org/10.1158/1078-0432.CCR-05-0964. [PubMed].
- 142. KLF6 inhibits estrogen receptor-mediated cell growth in breast cancer via a c-Src-mediated pathway. Molecular and Cellular Biochemistry. 2010; 335: 29-35. https://doi.org/10.1007/s11010-009-0237-8. [PubMed].
- 143. Quantification and clinical relevance of gene amplification at chromosome 17q12-q21 in human epidermal growth factor receptor 2-amplified breast cancers. Breast Cancer Research. 2011; 13: R15. https://doi.org/10.1186/bcr2824. [PubMed].
- 144. uPAR and HER2 Genes Are Usually Co-Amplified in Individual Breast Cancer Cells from Blood and Tissues. Breast Care (Basel). 2008; 3: 16–19. https://doi.org/10.1159/000151738. [PubMed].
- 145. The identification and characterization of breast cancer CTCs competent for brain metastasis. Science Translational Medicine. 2013; 5. https://doi.org/10.1126/scitranslmed.3005109. [PubMed].
- 146. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell. 2014; 158: 1110-1122. https://doi.org/10.1016/j.cell.2014.07.013. [PubMed].
- 147. Tumor Marker Utility Grading System: a Framework to Evaluate Clinical Utility of Tumor Markers. Journal of the National Cancer Institute. 1996; 88: 1456-1466. https://doi.org/10.1093/jnci/88.20.1456. [PubMed].
- 148. Thomssen C and Jänicke F. Do we need better prognostic factors in node-negative breast cancer? European Journal of Cancer. 2000; 36: 293-298. https://doi.org/10.1016/s0959-8049(99)00304-4. [PubMed].
- 149. Urokinase plasminogen activator and urokinase plasminogen activator receptor in breast cancer. International Journal of Cancer. 1995; 61: 597-600. https://doi.org/10.4103/0973-1482.174547. [PubMed].
- 150. Tumor stroma is the predominant uPA-, uPAR-, PAI-1-expressing tissue in human breast cancer: prognostic impact. Histology and Histopathology. 2009; 24: 869-877. https://doi.org/10.14670/HH-24.869. [PubMed].
- 151. Harbeck N, Ross JS, Yurdseven S, Dettmar P, Pölcher M, Kuhn W, Ulm K, Graeff H, and Schmitt M. HER-2/neu gene amplification by fluorescence in situ hybridization allows risk-group assessment in node-negative breast cancer. International Journal of Oncology. 1999; 14: 663-734. https://doi.org/10.3892/ijo.14.4.663. [PubMed].
- 152. HER-2/neu and Urokinase-Type Plasminogen Activator and Its Inhibitor in Breast Cancer. Clinical Cancer Research. 2001; 7: 2448-2457. [PubMed].
- 153. Jänicke F, Schmitt M, Pache L, Ulm K, Harbeck N, Höfler H, and Graeff H. Urokinase (uPA) and its inhibitor PAI-1 are strong and independent prognostic factors in node-negative breast cancer. Breast Cancer Research and Treatment. 1993; 24: 195-208. https://doi.org/10.1007/BF01833260. [PubMed].
- 154. Look MP, van Putten WLJ, Duffy MJ, Harbeck N, Christensen IbJ, Thomssen C, Kates R, Spyratos F, Fernö M, Eppenberger-Castori S, Sweep CGJF, Ulm K, Peyrat J-P, et al. Pooled Analysis of Prognostic Impact of Urokinase-Type Plasminogen Activator and Its Inhibitor PAI-1 in 8377 Breast Cancer Patients. Journal of the National Cancer Institute. 2002; 94: 116-128.
- 155. Abstract P6-06-44: Role of uPA and PAI-1 mRNA expression as prognostic factors in molecular subtypes of breast cancer. Cancer Research. 2013; 73: P6-06-44. https://doi.org/10.2147/OTT.S65344. [PubMed].
- 156. uPA and PAI-1 as biomarkers in breast cancer: validated for clinical use in level-of-evidence-1 studies. Breast Cancer Research. 2014; 16: 428. https://doi.org/10.1186/s13058-014-0428-4. [PubMed].
- 157. RNAi-mediated downregulation of UPAR synergizes with targeting of HER2 through the ERK pathway in breast cancer cells. International Journal of Cancer. 2010; 127: 1507-1516. https://doi.org/10.1002/ijc.25159. [PubMed].
- 158. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977; 197: 893-895. https://doi.org/10.1126/science.887927. [PubMed].
- 159. Giavazzi R, Alessandri G, Spreafico F, Garattini S, and Mantovani A. Metastasizing capacity of tumor cells from spontaneous metastases of transplanted murine tumors. British Journal of Cancer 1980; 42: 462-472. https://doi.org/10.1038/bjc.1980.259. [PubMed].
- 160. Mantovani A, Giavazzi R, Alessandri G, Spreafico F, and Garattini S. Characterization of tumor lines derived from spontaneous metastases of a transplanted murine sarcoma. European Journal of Cancer 1981; 17: 71-76. https://doi.org/10.1016/0014-2964(81)90213-9. [PubMed].
- 161. Milas L, Peters LJ, and Ito H. Spontaneous metastasis: random or selective? Clinical and Experimental Metastasis. 1983; 1: 309-315. https://doi.org/10.1007/BF00121193. [PubMed].
- 162. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002; 415: 530-536. https://doi.org/10.1038/415530a. [PubMed].
- 163. van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, et al. A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine. 2002; 347: 1999-2009. https://doi.org/10.1056/NEJMoa021967. [PubMed].
- 164. A molecular signature of metastasis in primary solid tumors. Nature Genetics. 2003; 33: 49-54. https://doi.org/10.1038/ng1060. [PubMed].
- 165. A progression puzzle. Nature. 2002; 418: 823. https://doi.org/10.1038/418823a. [PubMed].
Last Modified: 2016-06-03 06:50:31 EDT
